Research Article - International Journal of Medical Research & Health Sciences ( 2023) Volume 12, Issue 8
Differentiation of Breast Cancer Immunohistochemical Status Using Digital Mammography Radiomics Features
Malomon Aimé Bonou1,2*, Zouhour Ben Azouz2, Khlifa Nawres3, Rodrigue Sètchéou Allodji4 and Julien Dossou12Signals and Smart Systems Laboratory, National Engineering School of Tunis, University of Tunis El Manar, Tunisia
3Laboratory of Biophysics and Medical Technologies, Higher Institute of Medical Technology, 1006 Tunis, University of Tunis El Manar, Tunisia
4Cancer and Radiation, Unit 1018 INSERM, University Paris-Saclay, Gustave Roussy, 39, Rue Camille Desmoulins, 94805, Villejuif Cedex, France
Malomon Aimé Bonou, Non-Communicable Diseases and Cancer Research Unit, Laboratory of Applied Biology Research, Polytechnic School of Abomey-Calavi, University of Abomey-Calavi, Abomey-Calavi, Benin, Email: malombonou@yahoo.fr
Received: 25-Jul-2023, Manuscript No. ijmrhs-23-106296; Editor assigned: 27-Jul-2023, Pre QC No. ijmrhs-23-106296(PQ); Reviewed: 05-Aug-2023, QC No. ijmrhs-23-106296(Q); Revised: 08-Aug-2023, Manuscript No. ijmrhs-23-106296(R); Published: 30-Aug-2023
Abstract
Purpose: Discriminating breast cancer Hormonal Receptor (HR), human epidermal growth factor receptor (Her2) and Triple Negative (TN) status using mammography radiomic features. Materials and Methods: We used an open-source database enrolling 71 patients with confirmed breast cancer. It includes bilateral mammograms Craniocaudal (CC) and Mediolateral Oblique (MLO) as well as the breast cancer molecular status such as HR, Her2 and TN. We extracted a set of 486 quantitative descriptors from the original and the wavelets of the CC and the MLO mammograms. Using the training set (ntrain=48), we performed the features selection following two steps: (i) first, univariable feature selection had been implemented with correlation statistical test to eliminate redundancy between mammogram features. (ii) In second part, we used Support Vector Machine Recursive Feature Elimination (SVM-RFE) method in 10-folds Cross-Validation repeated 10 times. Also, we applied the Synthetic Minority Oversampling Technique to tackle the issue of imbalanced classes. After that, we carried out three binary molecular classification (HR vs non-HR, Her2 vs non-Her2, TN vs non-TN) using Logistic Regression. These classifications were performed using respectively CC and MLO features individually and in two combinations: sum “CC+MLO” and concatenation “CC and MLO”. After the validation step (ntest=17), Accuracy and Under Receiver Operating Characteristic curve (AUC) were adopted to assess the proposed model performance. Results: Accuracies and AUCs recorded for three molecular classes in validation step were respectively ranging from 0.69/0.75 to 0.88/0.90, 0.52/0.53 to 0.64/0.63 and 0.70/0.70 to 0.79/0.77 for Her2, HR, TN. The best performances achieved for HR and Her2 classification were CC image features and “CC and MLO” features for TN. There is a strong representation of wavelets features based in the features selected. Conclusion: Our results suggest that mammographic quantitative features especially wavelet-based could be used to differentiate the breast cancer molecular subtype
Keywords
Breast cancer, Radiomics, Mammogram, Immunohistochemical status
Introduction
Quantitative imaging or radiomics, based on quantitative analysis of image textures is a new application in clinical oncology [1]. For instance, Computer Aid Diagnosis (CAD) systems designed with this approach have proved their ability to improve cancer diagnosis from different medical imaging modalities. Moreover, recent studies revealed that radiomics can potentiality achieve a molecular subtyping than goes beyond uniquely discriminating a tumor as malignant or benign [2,3]. This information is of a particular interest for the breast cancer treatment which is known as an example of heterogenous cancer group with a complex biological behavior and a great clinical variability [4,5]. Based on a standard Immuno Histo-Chemistry (IHC) directed at cellular markers that reflect the availability of targeted therapies, breast cancer can be classified in three main groups: (a) hormone sensitive (ER or PR positive), (b) HER2 positive, sensitive to trastuzumab or (c) Triple Negative Breast Cancer (TNBC). This later is defined by the absence of ER, PR and HER2 amplification [6]. Molecular subtyping is beneficial for the diagnosis and the individualized treatment of breast cancers. Breast cancer molecular subtyping can also be performed using genic analysis by genetic analysis. However this procedure is expensive, and requires specialized equipment and technical expertise [7]. Thus, IHC is the most used for breast cancer molecular subtyping but it remains invasive. It requires tissue specimens that are typically extracted by a needle biopsy. Moreover, the biopsy sample corresponds to a part of the tumor. Hence, it provides partial information on its complete parenchyma. To the contrary, medical images yield a global view of the tumor heterogeneity. This medical imaging advantage is the fundamental reason for the advent of radiomics. It is based on the hypothesis that, quantitative texture features extracted from tumor and microenvironment images could predict its biology character [8]. Guided by the goal of proving a correlation between radiomic features and breast cancer molecular subtyping a few studies have used DCE-MRI (Dynamic Contrast Enhancement-Magnetic Resonance Imaging) [9,10]. The authors of these studies have chosen this imaging modality because it provides information on tumor morphology as well as its physiology. However, other modalities like ultrasonography, considered as morphologic imaging have been used to subtype breast cancer [11,12]. Also, mammography radiomic features have been recently used to subtype breast cancer molecular status [13,14]. In these last studies, features were extracted only from the original image while Ergin et al. reported that features from wavelets decomposition reveal better mammogram pattern. According to this study, wavelets features contribute more than the original image features in the predictive model building [15].
Mammography and ultrasonography are more accessible than DCE-MRI in developing countries and are frequently used in the diagnosis process. Unlike mammography ultrasonography is operator dependent. For this reason, mammography is an imaging modality that can be used easily in developing countries to practice radiomics. Hence, we have initiated this study to further explore the possibility of predicting of breast cancer molecular status from mammograms using features based on Image Biomarker Standardization Initiative (IBSI) recommendations [16]. Unlike the previous studies, we used features extracted from original image and its wavelet decomposition. Furthermore, for comparison purpose, we have used two combinations methods of mammograms images views descriptors.
Material and Methods
Patient’s Data
To achieve our goal we conducted a retrospective study using an open source data downloaded from figshare repository [17]. These anonymized data were collected and used within a study that was approved by the institutional review board. It aims to establish an association between digital mammography radiomic and breast cancer OncotypeDX and PAM50 recurrence scores. The study englobes a total of 71 breast cases with clinicopathologic information (age, TNM grading, ER, PR, and HER2 status), digital mammograms (cranial caudal CC and medio-lateral oblique MLO), microarray data and tumor segmentation on mammograms images. A digital mammography system (Selenia, Hologic, Bedford, MA), with an automatic intensity adjustment was used to acquire mammogram of 70 microns per pixel and 12-bits grayscale for codification. Manuel segmentation of tumors were performed by an experienced breast radiologist [18]. Six patients were excluded because either their molecular statuses are missing or they belongs two molecular statuses. Amongst the 65 patients of our cohort (n=35) were Hormonal Receptor positive (HR), (n=13) had the Human Epidermal Grown Factor Receptor 2 positive status (Her2), and (n=17) were Triple Negative (TN) with respective average age of 52 years, 50 years and 50 years. Their breast tumor characteristic is described in Table 1.
| Qualitatives descriptors | Her2 | HR | TN | |
|---|---|---|---|---|
| Age (year) | 50.38 (6.86) | 52.14 (10.69) | 50.53 (10.69) | |
| Size (mm) | 45.08 (25.59) | 39.23 (18.27) | 54.76 (42.34) | |
| Microcalfication | NEG | 7 (9.23) | 18 (26.15) | 9 (12.30) |
| POS | 6 (10.76) | 17 (27.69) | 8 (13.84) | |
| Menopausal Status | Post-Menopausal | 7 (10.76) | 20 (30.76) | 9 (13.84) |
| Pre-Menopausal | 6 (9.23) | 15 (23.07) | 8 (12.30) | |
| Circumscribed | NEG | 10 (15.34) | 28 (43.07) | 12 (18.46) |
| POS | 3 (4.61) | 7 (10.67) | 5 (7.69) | |
Radiomic Features Extraction
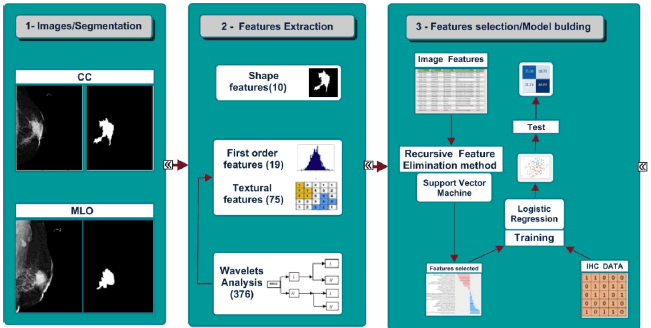
Dicom Mammograms and tumor segmentation images were decompressed with the open source Dicom viewer software MicroDicom 2.7.9. Tumor segmented images were rescaled between 0 and 1 grayscale with the python package ITKsimple [19]. The tumor region segmented from each mammogram view was used to extract 486 features as summarized in Figure 1. These features were divided into four types: shape features, first-order statistics, textural features and “Coiflet 1” wavelet decomposition-based features. All features extraction tasks have been performed using the python radiomics package, Pyradiomics 2 .1.2 [20].
Molecular Subtypes Classification
To assess the ability of predicting breast cancer molecular status from mammograms, we performed three binary classification tasks following in a strategy “a class against other classes” strategy: TN vs non-TN (HR and Her2),Her2 vs non-Her2 (HR and TN) and HR vs non-HR (Her2 and TN). Because of the large variation between features values, we performed a standard normalization following this formula:
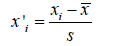
Where,x'i : normalized value of xi ,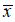 : mean. As the second pre-processing task, we applied the Synthetic Minority Oversampling Technique to tackle the issue of class imbalance specifically in TN vs non-TN and Her2 vs non-Her2 cases [21]. This method was performed in the same context by Ma et al, 2019 [13]. With the training dataset (ntrain=48) which represents 75% of all dataset, we implemented features selection in two steps. In the first part, we used the statistical correlation test to eliminate the redundancy between the mammogram features. Features with high correlation are more linearly dependent and hence have almost the same effect on the target variable. So, when two features have correlation coefficient value more than 0.8, we eliminated randomly one of two features. In second part, we used Support Vector Machine Recursive Feature Elimination (SVM-RFE) method in 10-folds Cross-Validation repeated 10 times and we plotted a graph showing the different cross-validation score (accuracy) in function of features number selected (Figure 2). SVM-RFE was introduced by Guyon et al. for selecting genes from microarray data analysis for cancer classification [22]. It includes four steps:
: mean. As the second pre-processing task, we applied the Synthetic Minority Oversampling Technique to tackle the issue of class imbalance specifically in TN vs non-TN and Her2 vs non-Her2 cases [21]. This method was performed in the same context by Ma et al, 2019 [13]. With the training dataset (ntrain=48) which represents 75% of all dataset, we implemented features selection in two steps. In the first part, we used the statistical correlation test to eliminate the redundancy between the mammogram features. Features with high correlation are more linearly dependent and hence have almost the same effect on the target variable. So, when two features have correlation coefficient value more than 0.8, we eliminated randomly one of two features. In second part, we used Support Vector Machine Recursive Feature Elimination (SVM-RFE) method in 10-folds Cross-Validation repeated 10 times and we plotted a graph showing the different cross-validation score (accuracy) in function of features number selected (Figure 2). SVM-RFE was introduced by Guyon et al. for selecting genes from microarray data analysis for cancer classification [22]. It includes four steps:
• Train an SVM on the training set;
• Calculate ranking criteria based on the SVM weights;
• Eliminate features with the smallest ranking criterion;
• Repeat the process.
SVM-RFE was also used in similarly studies for feature selection and it seemed very efficient in the context of small size dataset [23,24]. After feature selection step, we built predictive model using logistic regression classifier on the training set. All models had been validated on the test dataset (ntest=17). We used for each classification task four different images features from CC, MLO, CC+MLO, CC and MLO (features concatenation). All machine learning tasks were accomplished using the scikit-learn 0.20.3 of python 3 software [25].
Statistical Analysis
Descriptive statistic parameters are performed to summarize patient’s clinical information and their breast cancer qualitative characteristic (Table 1). Accuracy and Area Under receiver operating characteristic Curve (AUC) were used to evaluate the predictive performances of all classification models.
Result
Radiomics Tasks: Breast Cancer Molecular Classification Using Mammogram Quantitative Features
In the training step, we recorded for the three binary classifications using our database of mammogram features the following performances:
• Between 0.98/0.98 and 1/1 for the TN vs non-TN classification,
• Between 0.89/0.88 and 1/1 for the Her2 vs non-Her2,
• And from 0.93/0.93 to 1/1 for HR vs non-HR classification (Table 2).
| Mammograms features | CC | MLO | CON | SOM | |
|---|---|---|---|---|---|
| Molecular Status | TN | 0.98/0.98 | 0.98/0.98 | 1/1 | 0.97/0.97 |
| Her2 | 0.89/0.88 | 0.97/0.97 | 1/1 | 0.96/0.95 | |
| RH | 0.93/0.93 | 0.70/0.70 | 1/1 | 0.99/0.98 | |
Table 3 shows the results of the three binary differentiation of HR, Her2 and TN status.
| Mammograms features | CC | MLO | CON | SOM | |
|---|---|---|---|---|---|
| Molecular Status | TN | 0.75/0.74 | 0.75/0.73 | >0.79/0.77 | 0.70/0.70 |
| Her2 | 0.88/0.90 | 0.69/0.75 | 0.76/0.81 | 0.8/0.84 | |
| RH | 0.64/0.63 | 0.52/0.56 | 0.58/0.56 | 0.52/0.53 | |
Accuracies and AUCs achieved for the HR vs non-HR were between 0.52/0.53 and 0.64/0.63. The highest performance was provided by CC mammogram features.
In terms of the Her2 vs non-Her2 classification, performances ranges from 0.69/0.75 to 0.88/0.90. The best performance is achieved using CC image feature.
In the TN differentiation from other molecular classes, the achieved performances are between 0.70/0.70 and 0.79/0.77. “CC and MLO” features allowed achieving the highest performance for TN molecular status differentiation from other molecular status.
After the different molecular status classification task performed, mammogram features allowed us to differentiate better Her2 and TN breast cancer than HR breast cancer. Mammogram feature got from CC and MLO images combination were not systematically contributed to the classification performance improvement. Among six (06) binary classifications performed with “CC+MLO” and “CC and MLO” features, only in case of TN breast cancer differentiation “CC and MLO” features contributed to the highest classification performance.
Features Contribution
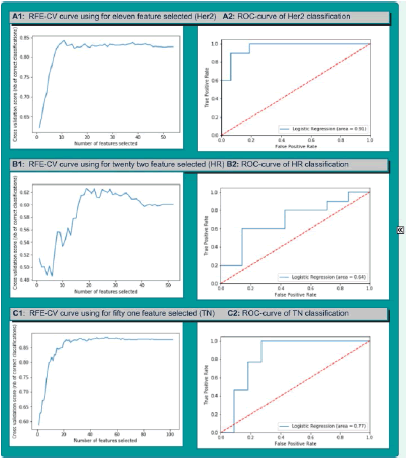
According to the achieved performances, wavelet decomposition features are more represented in feature selected than shape, first order and textural features. In case of highest performances there are 15 wavelet features over 22 features selected for HR breast cancer classification, 36 wavelet features over 51 features selected for the TN breast cancer classification and 5 wavelet features over 11 features selected for the Her2 breast cancer classification (Table 4, Figure 2).
| Features | HR vs non-HR | Her2 vs non-Her2 | TN vs non-TN |
|---|---|---|---|
| 0.64/0.63 | 0.88/0.90 | 0.79/0.77 | |
| Shape | 1 | 1 | 3 |
| 1st order | 4 | 1 | 8 |
| Textural | 2 | 4 | 4 |
| Wavelet | 15 | 5 | 36 |
| Total | 22 | 11 | 51 |
Discussion
Identifying the Hormonal Receptor (HR) and human epidermal growth factor receptor (Her2) status is valuable for an adequate therapeutic choice [26]. This information has been for a long time considered as exclusive to biological tests. However, recent studies has accumulated evidence that medical images can potentially provide this information [9,11,13,14,27,28].
In this radiomic study, we have attempted to predict breast cancer molecular status using only mammogram quantitative features. For this aim, we carried three binary of breast cancer subtypes classification using separately mammogram views descriptors (CC, MLO) and combined mammogram views descriptors (“CC+MLO”, “CC and MLO”). Among all binary classifications, we recorded the highest classification performance of Her2 and HR breast cancer using features extracted from a single view. As for the TN classification image combined features (“CC and MLO”) view provided the highest performance. Our results suggest that using features combination from both mammograms views could increase the model performance however their effect is not systematic.
About the best performance achieved using the logistic regression to distinguish each molecular class from the other was: Accuracy/AUC 0.79/0.77 for TN vs non-TN, Accuracy/AUC 0.88/0.90 for Her2 non-Her2 and 0.64/0.63for HR vs non-HR. In the case for Her2 prediction, our performance outperform that achieved by Ma et al, and Zhou et al. which were respectively 0.748/0.784 and 0.787 (AUC) [13,14]. About the TN breast cancer molecular status prediction, our highest performance recorded was close to that of Ma et al which was 0.796/0.865. Finally our prediction in the HR breast molecular was less good than what recorded by Ma et al (Accuracy/AUC: 0.788/0.752) [13]. Textural radiomics features used in these previous studies were limited to gray-level cooccurrence matrix (GLCM), Grey-Level Run Length Matrix (GLRLM) and, Grey-Level Size Zone Matrix (GLSZM) features from original images in case Zhou et al. study and to Gray-Level Cooccurrence Matrix (GLCM) in the study of Ma et al. [13,14]. In our study we used more features that get us an advantage to capture the difference between breast molecular subtypes, but our small study sample was a disadvantage relative to previous studies.
Among the best features ranked and selected for the radiomic task, we have noticed that a better representation of features extracted from wavelets decomposition. This finding has already been made by previous study reporting that wavelet transform seems to bring out mammograms patterns of texture [18].Yang et al. found a large representation of wavelets features in their mammography radiomic features selected for the axillary lymph node metastasis prediction [29]. These findings show the importance to include the wavelets features in mammography radiomic task.
According to the materiel and methodology use, this study presents some limitations. The small sample size with an over-representation of HR class compared to other classes was the first limitation. All mammograms used, are acquired through the same mammography system. Third, the manual segmentation is a limit to real tumor delineation.
Conclusion
In this study we used only mammogram IBSI quantitative features to predict the molecular status of breast cancer. We recorded the moderate performance mostly for Her2 and TN breast cancer prediction and less good performance for the HR breast cancer. Also, it has been noticed a strongly participation of wavelets features in the highest classification performance. Our results suggest that mammographic quantitative features especially wavelet-based could be used to differentiate the breast cancer molecular subtype. A future large and multicentric study, including African data, will confirm and improve our observations. This perspective will allow getting a tool that could help pathologist or clinician for complementary analysis beside pathological exam.
Acknowledgment
We extend our heartfelt appreciation to the Entreprenariat, Ressources, Management, Innovation et Technologies (ERMIT) program for the invaluable support of funded mobility. Their assistance has not only broadened our horizons but also significantly contributed to our academic and professional development. The ERMIT program's dedication to promoting innovation and entrepreneurship is commendable, and we are grateful for the opportunity to connect with experts and peers in our field. The financial support provided has alleviated financial constraints and allowed us to fully engage in our research and skill enhancement. Our sincere thanks to the ERMIT program for their instrumental role in this enriching experience.
Declarations
Conflict of Interest
The authors declared no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
References
- Yip, Stephen SF, and Hugo JWL Aerts. "Applications and limitations of radiomics." Physics in Medicine & Biology, Vol. 61, No. 13, 2016, p. 150.
Google Scholar Crossref - Crivelli, Paola, et al. "A new challenge for radiologists: radiomics in breast cancer." BioMed research international, 2018.
Google Scholar Crossref - Sanduleanu, Sebastian, et al. "Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score." Radiotherapy and Oncology, Vol. 127, No. 3, 2018, pp. 349-60.
Google Scholar Crossref - Uscanga-Perales, et al. "Triple negative breast cancer: Deciphering the biology and heterogeneity." Medicina universitaria, Vol. 18, No. 71, 2016, pp. 105-14.
Google Scholar Crossref - Esparza-López, José, et al. "Breast cancer intra-tumor heterogeneity: one tumor, different entities." Revista de investigacion clinica, Vol. 69, No. 2, 2017, pp. 66-76
. Google Scholar - Goldhirsch, Aron, et al. "Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013." Annals of oncology, Vol. 24, No. 9, 2013, pp. 2206-23.
Google Scholar Crossref - Dai, Xiaofeng, Ana Chen, and Zhonghu Bai. "Integrative investigation on breast cancer in ER, PR and HER2-defined subgroups using mRNA and miRNA expression profiling." Scientific reports, Vol. 4, No. 1, 2014, p. 6566.
Google Scholar Crossref - Sutton, Elizabeth J., et al. "Breast cancer subtype intertumor heterogeneity: MRIâ?based features predict results of a genomic assay." Journal of Magnetic Resonance Imaging, Vol. 42, No. 5, 2015, pp. 1398-406.
Google Scholar Crossref - Monti, Serena, et al. "DCE-MRI pharmacokinetic-based phenotyping of invasive ductal carcinoma: a radiomic study for prediction of histological outcomes." Contrast Media & Molecular Imaging, 2018.
Google Scholar Crossref - Agner, Shannon C., et al. "Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study." Radiology, Vol. 272, No. 1, 2014, pp. 91-9.
Google Scholar Crossref - Guo, Yi, et al. "Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma." Clinical breast cancer, Vol. 18, No. 3, 2018, pp. 335-44.
Google Scholar Crossref - Lee, Si Eun, et al. "Radiomics of US texture features in differential diagnosis between triple-negative breast cancer and fibroadenoma." Scientific reports, Vol. 8, No. 1, 2018, pp. 1-8.
Google Scholar Crossref - Ma, Wenjuan, et al. "Breast cancer molecular subtype prediction by mammographic radiomic features." Academic radiology, Vol. 26, No. 2, 2019, pp. 196-201.
Google Scholar Crossref - Zhou, Jing, et al. "Evaluating the HER-2 status of breast cancer using mammography radiomics features." European Journal of Radiology, Vol. 121, 2019, p. 108718.
Google Scholar Crossref - Ergin, Semih, and Onur Kilinc. "A new feature extraction framework based on wavelets for breast cancer diagnosis." Computers in biology and medicine, Vol. 51, 2014, pp. 171-82.
Google Scholar Crossref - Zwanenburg, A., S. Leger, and M. Vallières and Lock S. "Image biomarker standardisation initiative." arXiv preprint, 2019.
Google Scholar Crossref - Trevino V. Breast Cancer Images & Segmentation - Correlation of Gene Expression Subtypes and Image Features. Figshare, 2018.
- Tamez-Pena, Jose-Gerardo, et al. "Radiogenomics analysis identifies correlations of digital mammography with clinical molecular signatures in breast cancer." PloS one, Vol. 13, No. 3, 2018.
Google Scholar Crossref - Yaniv, Ziv, et al. "SimpleITK image-analysis notebooks: a collaborative environment for education and reproducible research." Journal of digital imaging, Vol. 31, No. 3, 2018. pp. 290-303.
Google Scholar Crossref - Van Griethuysen, Joost JM, et al. "Computational radiomics system to decode the radiographic phenotype." Cancer research, Vol. 77, No. 21, 2017, pp. 104-7.
Google Scholar Crossref - Chawla, Nitesh V., et al. "SMOTE: synthetic minority over-sampling technique." Journal of artificial intelligence research, Vol. 16, 2002, pp. 321-57.
Google Scholar Crossref - Guyon, Isabelle, et al. "Gene selection for cancer classification using support vector machines." Machine learning, Vol. 46, 2002, pp. 389-422.
Google Scholar Crossref - Vabalas, Andrius, et al. "Machine learning algorithm validation with a limited sample size." PloS one, Vol. 14, No. 11, 2019.
Google Scholar Crossref - Zhang, Fan, et al. "Recursive SVM biomarker selection for early detection of breast cancer in peripheral blood." BMC medical genomics, Vol. 6, No. 1, 2013, pp. 1-10.
Google Scholar Crossref - Pedregosa, Fabian, et al. "Scikit-learn: Machine learning in Python." The Journal of machine Learning research, Vol. 12, 2011, pp, 2825-30.
Google Scholar - Bauer, Katrina R., et al. "Descriptive analysis of estrogen receptor (ER)â?negative, progesterone receptor (PR)â?negative, and HER2â?negative invasive breast cancer, the soâ?called tripleâ?negative phenotype: a populationâ?based study from the California cancer Registry." Cancer, Vol. 109, No. 9, 2007, pp. 1721-8.
Google Scholar Crossref - Samala, Ravi K., et al. "Identifying key radiogenomic associations between DCE-MRI and micro-RNA expressions for breast cancer." Medical Imaging 2017: Computer-Aided Diagnosis, Vol. 10134, 2017.
Google Scholar Crossref - Fan, Ming, et al. "Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer." PloS one, Vol. 12, No. 2, 2017.
Google Scholar Crossref - Yang, Jingbo, et al. "Preoperative prediction of axillary lymph node metastasis in breast cancer using mammography-based radiomics method." Scientific Reports, Vol. 9, No. 1, 2019, p. 4429.
Google Scholar Crossref