Research - International Journal of Medical Research & Health Sciences ( 2020) Volume 9, Issue 12
miR-4772-5p and miR-196b as Potential Prognostic Biomarker in Pediatric Core Binding Factor Acute Myeloid Leukemia
Vikas Gaur1,2, Shilpi Chaudhary1, Suyash Agarwal3,4, Surender K. Sharawat1, Sameer Bakhshi1, Pankaj Sharma2 and Sachin Kumar1*2Amity Institute of Biotechnology, Amity University, Uttar Pradesh, Noida, India
3ICMR Computational Genomics Centre, Indian Council of Medical Research, New Delhi, India
4Informatics, Systems and Research Management, Indian Council of Medical Research, New Delhi, India
Sachin Kumar, Department of Medical Oncology, Dr. B. R. A. Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, India, Tel: 011 2658 8500, Email: sksingla@aiims.ac.in, psharma8@amity.edu
Received: 19-Nov-2020 Accepted Date: Dec 21, 2020 ; Published: 28-Dec-2020
Abstract
Introduction: Aberrant expression of miRNAs has been linked with the initiation and progression of several cancers. However, their role as prognostic biomarkers in pediatric core binding factor AML (CBF-AML) is still unclear. The objective of the present study was to identify differentially expressed miRNAs and their prognostic significance in pediatric CBF-AML patients. Methods: Bone marrow samples from 28 pediatric CBF-AML patients and 27 pediatric controls were obtained after ethical approval. Based on our previous findings, expression levels of 11 miRNAs and their selected potential targets were analysed using TaqMan advanced miRNA assay and SYBR Green-based qRT-PCR, respectively. Differential miRNA expression was correlated with clinicopathological parameters and survival outcomes. Results: Upregulation of miR-100-5p, miR-4446-3p, and miR-335-3p and downregulation of miR-409-3p, miR-151a-3p, miR-196b, miR-4772-5p, and miR-758-3p was observed in pediatric CBF-AML patients as compared to controls. Low miR-196b expression was associated with poor disease-free and overall survival and low expression of miR-4772-5p was found to be an independent predictor of poor event-free survival. Except PAX-5, other predicted gene targets were found to be upregulated in pediatric CBF-AML patients. Conclusion: miR-196b and miR-4772-5p may be considered as independent prognostic biomarkers in pediatric CBF-AML. The miRNA-mRNA interaction network suggests potential involvement of miRNAs, which needs to be further, confirmed using mechanistic studies.
Keywords
MicroRNA, Acute myeloid leukemia, Survival, Cytogenetics, Prognosis, Biomarker
Introduction
Acute Myeloid Leukemia (AML) is a hematopoietic stem cell disorder characterised by expansion of undifferentiated myeloid progenitor cells [1,2]. The diagnosis and prognosis of AML is based on clinical symptoms, cell morphology, immunophenotyping, conventional and molecular cytogenetics, and mutation analysis [2]. Though largely affecting adults, AML is the second most common pediatric leukemia after Acute Lymphoblastic Leukemia (ALL) and accounts for 15-20% of all pediatric leukaemia’s. Intensive therapy and supportive care have improved the 5-year Overall Survival (OS) rates to 65-70%. However, duration of OS varies according to the AML subtypes and other influential factors [2]. Karyotype presentation at the time of diagnosis is the most important prognostic factor. Around 40-50% of adult and 25-30% of pediatric AML patients are cytogenetically normal [3], placed in the intermediate risk group and are treated accordingly. AML patients carrying t(8;21) or inv(16) cytogenetic abnormalities are classified as Core Binding Factor (CBF)-AML and they have a favourable disease outcome. There are, however, significant differences in disease diagnosis, prognosis and treatment responses for adult and pediatric AML that is increasingly being recognised.
Apart from various cytogenetic and genetic factors, few studies have also looked at the role of epigenetic factors, such as non-coding RNAs in leukemogenesis. MicroRNAs (miRNAs) are made up of highly conserved 18-25 nucleotides that do not code for any protein [4–6]. They are considered to be negative post-transcriptional regulators of gene expression that function by binding to an imprecisely complimentary 3’ Untranslated Region (UTR) of a target mRNA in the cytoplasm. However, increasing reports suggest that miRNAs in the nucleus may also promote gene expression by binding to cognate promoters [7]. Dysregulation in the expression of a specific or a group of miRNAs has been observed in various human diseases, including haematological malignancies [8]. The role of miRNAs in the regulation of diverse cellular and biological processes involved in the initiation and progression of cancer is also well elucidated [9,10].
A number of studies have found dysregulated miRNAs expression in both adult and pediatric AML patients. Using small RNA sequencing, we have recently identified 66 upregulated and 102 downregulated miRNAs in the bone marrow of pediatric Cytogenetically Normal AML (CN-AML) patients [11]. Further qRT-PCR-based validation of 11 miRNAs revealed a significant upregulation of miR-196b, miR-100-5p, miR-1275, miR-4446-3p, and miR-335-3p, while the expression of miR-151a-3p, miR-409-3p, miR-4772-5p, and miR-758-3p was significantly downregulated [11]. Interestingly, the expression of miR-4446-3p and miR-758-3p significantly correlated with survival outcome indicating their potential utility as prognostic biomarkers [11]. In the present study, we investigated if the expression of any of these 11 miRNAs (along with their putative target genes) is dysregulated in pediatric CBF-AML patients, and whether they have any prognostic significance. We also established miRNA-mRNA interaction network to identify perturbed molecular pathways in CBF-AML.
Materials and Methods
Patients
This study included 28 newly diagnosed and untreated patients of pediatric CBF-AML and 27 pediatric controls. All the subjects were recruited from the outpatient Department of Medical Oncology, All India Institute of Medical Sciences, New Delhi, India between 2016 and 2019. Pediatric patients with solid tumors without any Bone Marrow (BM) involvement were taken as controls. From every subject, demographic and baseline clinical data was collected. The diagnosis of AML was based on cellular morphology, cytochemistry, immunophenotyping and percentage of blast from BM aspirate and/or peripheral blood using flow cytometry, BM/blood smears, and cytogenetic analysis. All patients received 3+7 induction chemotherapy with daunorubicin (60mg/m or 90 mg/m on days 1, 2 and 3) and cytarabine (100 mg/m /daily for one week) followed by consolidation therapy after achievement of Complete Remission (CR). Patients older than 18 years previously treated with anti-leukemic therapy or not classified as CBF-AML were excluded from the study. The study was approved by the Institutional Ethics Committee and informed written consent was obtained from all the subjects.
Mutation Analysis
BMMCs were isolated by density gradient centrifugation using Histopaque 1077 (Thermo Fisher Scientific) from 5 mL of BM. The purified BMMCs were used for the isolation of genomic DNA and total RNA using PureLink Genomic DNA mini kit (Thermo Fisher Scientific) and TRIzol (Thermo Fisher Scientific), respectively. The quantification of RNA and DNA was performed by Nanodrop 1000 (Thermo Fisher Scientific) while their quality was assessed by agarose gel electrophoresis.
Genomic DNA was used for the identification of Fms-related tyrosine kinase 3 Internal Tandem Duplication (FLT3- ITD) and FLT3 Tyrosine Kinase Domain (TKD) mutations using Polymerase Chain Reaction (PCR). RNA was used for detecting the mutation in Nucleophosmin-1 (NPM1) gene using reverse transcriptase allele-specific oligonucleotide PCR [12,13]. No further mutation analysis could be undertaken due to limited availability of nucleic acid from patient samples.
Analysis of miRNA Expression by qRT-PCR
Based on the findings of our previous study in CN-AML patients, we selected 11 miRNAs for the analysis of their expression using TaqMan advanced miRNA assay [11]. Briefly, cDNA was prepared from total RNA using TaqMan advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific) as per recommended protocol. The miRNA-specific TaqMan advanced miRNA assays (Applied Biosystems) were used for the quantification of individual miRNAs on Roche Light Cycler 480 in compatible white 96-well plates (Roche) using TaqMan fast advanced master mix (Thermo Fisher Scientific). The relative expression of miRNAs was calculated by 2-ΔΔCt method after normalizing the expression of target miRNA with recommended endogenous miR-423.
Prediction of Potential Targets of miRNAs and Analysis of their Expression
The potential targets of selected miRNAs were predicted using miRanda v3.3a, with cut-off values, including score ≥140, complementarity ≥60%, alignment length ≥10 and total energy ≤-20. The expression of the predicted targets was checked by SYBR Green qRT-PCR assay (Thermo Fisher Scientific). Primers of target genes were designed using online tools such as primer blast and primer 3. The relative gene expression was calculated by 2-ΔΔCt method after normalizing target gene expression with the expression of β-actin.
Statistical Analysis
Complete remission was defined as <5% of blast in BM/blood. Event-Free Survival (EFS) was measured from the date of diagnosis to the death or relapse whichever comes first. Disease-Free Survival (DFS) was calculated from the date of complete remission to relapse or death, while OS was calculated from the date of diagnosis till the death or last follow up. The stratified log rank test was used for univariate comparison. All patients were grouped into quartiles according to miRNAs expression levels and divided into high and low based on the trend observed in the clinical outcomes after performing a Cox regression analysis of EFS, DFS and OS with all validated miRNA quartiles grouping as independent variables. The Kaplan Meier method was used to estimate the probability of EFS, DFS and OS. Multivariate analysis was performed using Cox regression model. Student’s t-test was used to determine the statistical significance of experimental results and a p<0.05 was considered as statistically significant. SPSS 23 and GraphPad prism 8 was used to process the obtained data.
Results
Patient Characteristics
The mean age of the pediatric CBF-AML and pediatric controls was 10.9 years and 10 years, respectively. Most of the patients were male (60.7%). Their median EFS, DFS and OS were 465 days, 446 days, and 547 days, respectively. The median BM blast percentage was 60% (range of 20%-95%). Almost 93% of the patients achieved CR after first induction and 12.5% of patients required re-induction. Consolidation therapy was given to all patients who achieved CR. None of the patient underwent allogenic hematopoietic stem cell transplantation. In the present study, the data on minimal residual diseases status of pediatric CBF-AML patients was not available. The clinico-pathological characteristics of CBF-AML patients are listed in Table 1. Out of 28 CBF-AML patients, 26 were having t(8;21) (q22;q22) while 2 were positive for inv(16) abnormality. The analysis of FLT3-ITD/TKD (n=21) and NPM1 (n=15) mutations indicated the presence of FLT3-ITD mutation in only one patient, while none of the patient carried NPM1 mutation. We could not perform any additional molecular analysis, including mutations in KIT gene due to unavailability of sample.
| Clinical parameters | Total patients | EFS | DFS | OS | |||
| Chi-Square | p-value | Chi-Square | p-value | Chi-Square | p-value | ||
| Age (years) | Median: 10.5 | 0.03 | 0.86 | 0.03 | 0.87 | 0.22 | 0.63 |
| Range:3-17 | |||||||
| Gender | Male: 17 (60.7%) | 2.04 | 0.15 | 1.69 | 0.19 | 0.17 | 0.68 |
| Female: 11(39.3%) | |||||||
| Total leukocyte count (mm3) | Median: 11270 | 0.04 | 0.83 | 0.05 | 0.82 | 0.12 | 0.72 |
| Range: 3260-320000 | |||||||
| Absolute neutrophil count (mm3) | Median: 5135 | 0.02 | 0.88 | 0.02 | 0.89 | 1.65 | 0.2 |
| Range: 590-186000 | |||||||
| Platelets (per microliter) | Median: 42000 | 0.03 | 0.87 | 0.05 | 0.82 | 3.35 | 0.07 |
| Range: 2600-190000 | |||||||
| Haemoglobin (g/dl) | Median: 7.6 | 0.19 | 0.66 | 0.44 | 0.5 | 0.07 | 0.78 |
| Range: 5.4-12.4 | |||||||
| Bone marrow blast (%) | Median: 60 | 3.36 | 0.07 | 3.65 | 0.08 | 0.87 | 0.35 |
| Range: 20-95 | |||||||
| Lactate dehydrogenase (U/L) | Median: 642 | 0.03 | 0.86 | 0.02 | 0.88 | 1.31 | 0.25 |
| Range: 247-2175 | |||||||
EFS: Event-Free Survival; DFS: Disease-Free Survival; OS: Overall Survival
Table 1 Clinico-pathological characteristics and their correlation with survival of pediatric CBF-AML patients
Differential Expression of miRNAs and Their Predicted Targets in Pediatric CBF-AML Patients
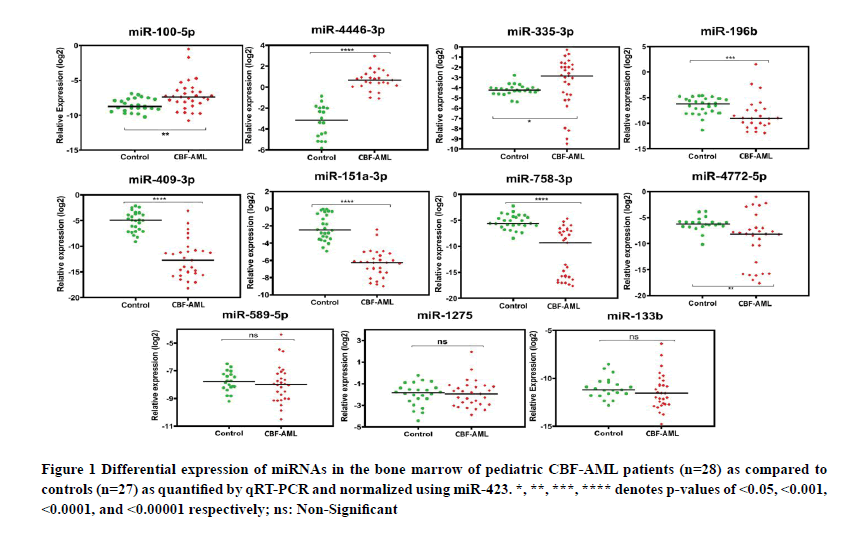
We found a significant upregulation of 3 miRNAs (miR-100-5p, miR-335-3p, and miR-4446-3p), while 5 miRNAs (miR-4772-5p, miR-758-3p, miR-151a-3p, miR-196b and miR-409-3p) were significantly downregulated in pediatric CBF-AML patients as compared to controls. In contrast, the expression levels of miR-133b, miR-1275, and miR- 589-5p remained largely unchanged (Figure 1). While miR-100 expression correlated significantly with Total Leukocyte Counts (TLC), none of the other differentially expressed miRNAs were found to have any correlation with any clinicopathological parameter tested, including age, gender, TLC, absolute neutrophil counts, platelet counts, haemoglobin levels, BM blast percentage and lactate dehydrogenase levels (Table 2).
| Clinical parameters (N) | miR-409-3p | miR-758-3p | miR-4772-5p | miR-4446-3p | miR-151a-3p | miR-100-5p | miR-196b | miR-335-3p | ||||||||
| median | p-value | median | p-value | median | p-value | median | p-value | median | p-value | median | p-value | median | p-value | median | p-value | |
| Age (years) | 0.16 | 0.055 | 0.09 | 0.42 | 0.28 | 0.9 | 0.61 | 0.74 | ||||||||
| <10 (14) | -9.87 | -9.94 | -3.48 | 4.16 | -4.02 | 1.09 | -2.24 | 1.2 | ||||||||
| >10 (14) | -6.3 | -2.27 | -1.44 | 3.71 | -4.42 | 10.8 | -2.49 | 1.36 | ||||||||
| Gender | 0.78 | 0.51 | 0.45 | 0.62 | 0.28 | 0.1 | 0.77 | 0.33 | ||||||||
| Male (17) | -6.21 | -1.86 | -1.06 | 4.08 | -3.91 | 1.66 | -2.26 | 1.56 | ||||||||
| Female (11) | -8.86 | -8.7 | -1.96 | 3.94 | -4.68 | 0.8 | -2.25 | 0.73 | ||||||||
| TLC (mm3) | 0.87 | 0.87 | 0.19 | 0.49 | 0.76 | 0.02 | 0.17 | 0.16 | ||||||||
| ≤11,270 (14) | -7.99 | -6.15 | -1.98 | 4.09 | -4.02 | 0.5 | -2.57 | 1.93 | ||||||||
| >11,270 (14) | -8.52 | -6.47 | -1.09 | 3.8 | -4.72 | 1.5 | -2.02 | 0.61 | ||||||||
| ANC (mm3) | 0.54 | 0.99 | 0.33 | 0.6 | 0.57 | 0.31 | 0.222 | 0.17 | ||||||||
| ≤5135 (13) | -8.86 | -3.53 | -1.95 | 4.09 | -4.17 | 0.76 | -2.51 | 1.88 | ||||||||
| >5135 (13) | -7.57 | -9.42 | -0.77 | 3.73 | -3.91 | 1.29 | -1.45 | 0.5 | ||||||||
| Platelets (/uL) | 0.42 | 0.51 | 0.51 | 0.19 | 0.37 | 0.45 | 0.79 | 0.95 | ||||||||
| ≤42,000 (14) | -8.91 | -3 | -1.59 | 3.89 | -4.68 | 0.91 | -2.87 | 1.23 | ||||||||
| >42,000 (14) | -8.5 | -9.29 | -2.42 | 4.1 | -3.94 | 1.24 | -2.42 | 1.15 | ||||||||
| Hb (g/dl) | 0.79 | 0.94 | 0.4 | 0.31 | 0.13 | 0.28 | 0.34 | 0.23 | ||||||||
| ≤8 (18) | -8.91 | -6.15 | -2.12 | 3.84 | -4.12 | 1.19 | -2.53 | 1.13 | ||||||||
| >8 (10) | -6.75 | -6.47 | -1.59 | 4.14 | -3.86 | 0.8 | -1.64 | 1.68 | ||||||||
| BM blast (%) | 0.11 | 0.96 | 0.89 | 0.08 | 0.29 | 0.86 | 0.43 | 0.21 | ||||||||
| ≤60 (15) | -9.46 | -8.26 | -1.91 | 3.45 | -4.06 | 1.03 | -2.87 | 1.1 | ||||||||
| >60 (13) | -6.06 | -3.53 | -1.96 | 4.1 | -3.97 | 1.24 | -2.42 | 1.97 | ||||||||
| LDH (U/L) | 0.52 | 0.48 | 0.78 | 0.57 | 0.31 | 0.92 | 0.6 | 0.53 | ||||||||
| ≤642 (12) | -7.99 | -6.11 | -1.65 | 4.09 | -4.12 | 0.8 | -0.93 | 1.2 | ||||||||
| <642 (11) | -9.46 | -10.3 | -1.42 | 3.89 | -3.79 | 1.24 | -2.48 | 1.16 | ||||||||
TLC: Total Leukocyte Counts; ANC: Absolute Neutrophil Counts; Hb: Haemoglobin; BM: Bone Marrow; LDH: Lactate Dehydrogenase
Table 2: Clinico-pathological characteristics and their correlation with miRNA expression of pediatric CBF-AML
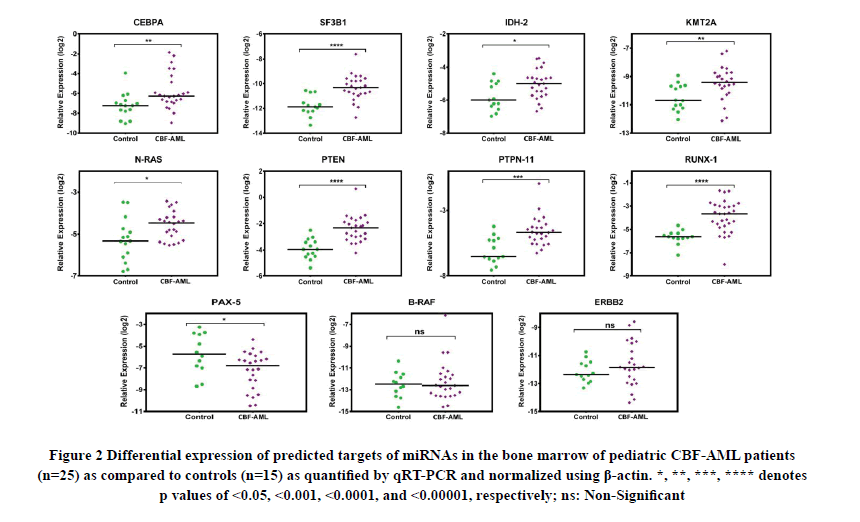
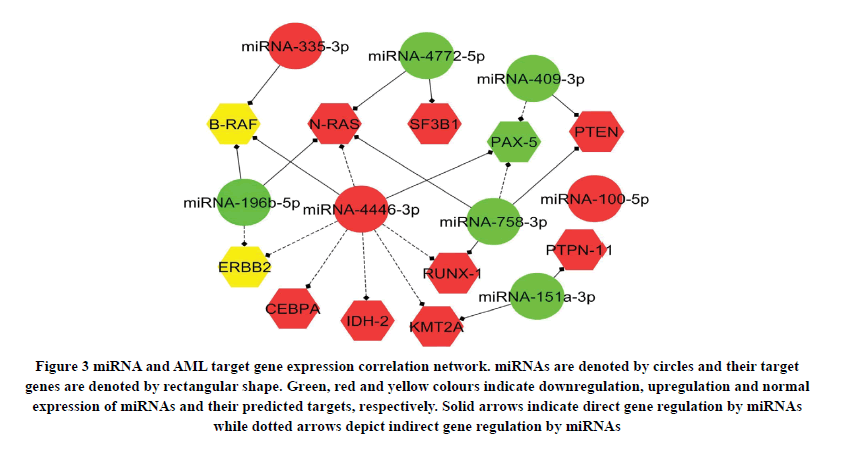
For analysing the expression of predicted targets of differentially expressed miRNAs, 11 genes were selected based on their involvement in the pathogenesis of AML, with a preference for genes targeted by multiple miRNAs. While we found a significant upregulation in the expression of 8 target genes (CEBPA, SF3B1, IDH-2, KMT2A, N-RAS, PTEN, PTPN-11 and RUNX-1), PAX-5 was downregulated in pediatric CBF-AML patients as compared to controls (Figure 2). Network analysis revealed that miRNAs may regulate their predicted targets either directly or indirectly (Figure 3). For example, miR-4446-3p can directly regulate PAX-5 while indirectly regulating N-RAS, CEBPA, IDH-2, KMT2A, and RUNX-1 (Figure 3).
Figure 2. Differential expression of predicted targets of miRNAs in the bone marrow of pediatric CBF-AML patients (n=25) as compared to controls (n=15) as quantified by qRT-PCR and normalized using β-actin. *, **, ***, **** denotes p values of <0.05, <0.001, <0.0001, and <0.00001, respectively; ns: Non-Significant
Figure 3. miRNA and AML target gene expression correlation network. miRNAs are denoted by circles and their target genes are denoted by rectangular shape. Green, red and yellow colours indicate downregulation, upregulation and normal expression of miRNAs and their predicted targets, respectively. Solid arrows indicate direct gene regulation by miRNAs while dotted arrows depict indirect gene regulation by miRNAs
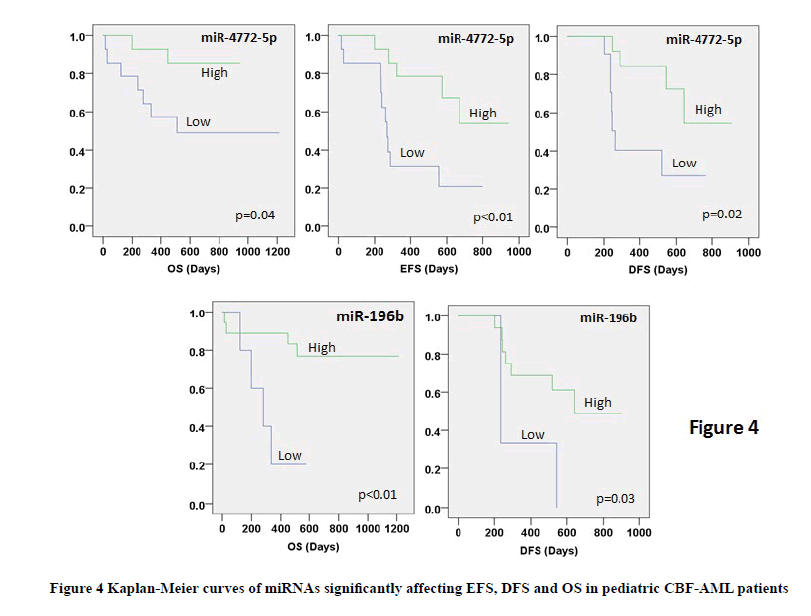
Correlation of miRNA Expression with Survival Outcomes in CBF-AML Patients
Lower expression of miR-4772-5p was significantly associated with poor EFS, DFS, and OS. Similarly, lower expression of miR-196b was significantly associated with poor DFS and OS in CBF-AML patients (Table 3, Figure 4). None of the remaining miRNAs as well as clinico-pathological characteristics correlated with any survival outcome (Table 3). Multivariate analysis (after adjustment for age and gender) revealed miR-4772-5p to be an independent prognostic factor for EFS while miR-196b was found to be independently associated with DFS and OS in pediatric CBF-AML patients (Table 4).
| S. No. | miRNAs | Expression profile | EFS (Days) | DFS (Days) | OS (Days) | |||
|---|---|---|---|---|---|---|---|---|
| Median (95% CI) | p-value | Median (95% CI) | p-value | Median (95% CI) | p-value | |||
| 1. | miR-409-3p | Low (15) | 671 (220.71-1121.28) | 0.62 | 642 (128.8-1155.1) | 0.55 | Not reached | 0.83 |
| High(13) | 574 (121.09-1026.99) | Not reached | ||||||
| 2. | miR-758-3p | Low(15) | 325 (0-846.35) | 0.98 | 642 (0-1416.67) | 0.94 | Not reached | 0.92 |
| High(13) | 556 (206.12-905.87) | 545 (487.2-602.7) | ||||||
| 3. | miR-4772-5p | Low(14) | 271(231.53-310.46) | <0.01 | 261 (232.34-289.65) | 0.02 | 511(Not reached) Not reached |
0.04 |
| High(14) | Not reached | Not reached | ||||||
| 4. | miR-4446-3p | Low(12) | 671 (Not reached) | 0.46 | Not reached | 0.07 | Not reached | 0.55 |
| High(12) | 325 (81.35-568.64) | 292(49.23-534.76) | ||||||
| 5. | miR-151a-3p | Low(15) | 574(80.14-1067.86) | 0.82 | (Not reached) | 0.78 | Not reached | 0.6 |
| High(13) | 556 (171.25-940.74) | 642 (235.7-1048.3) | ||||||
| 6. | miR-100-5p | Low(14) | 556 (140.23-971.76) | 0.62 | 521 (218.3-823.7) | 0.35 | Not reached | 0.57 |
| High(13) | 574 (Not reached) | Not reached | ||||||
| 7. | miR-196b | Low(5) | 263(192.44-333.56) | 0.053 | 235 (Not reached) | 0.03 | 281 (104.94-457.06) Not reached |
<0.01 |
| High(18) | 671 (Not reached) | 642(Not reached) | ||||||
| 8. | miR-335-3p | Low(15) | 280 (0-585.99) | 0.13 | 545 (0-1206.79) | 0.22 | Not reached | 0.8 |
| High(13) | Not reached | Not reached | ||||||
EFS: Event-Free Survival; DFS: Disease-Free Survival; OS: Overall Survival; CI: Confidence Interval
Table 3 Univariate analysis of miRNA expression with survival outcomes in pediatric CBF-AML patients
| Variables | EFS | DFS | OS | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p -value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| miR-4772-5p | 0.249 (0.078-0.798) | 0.019 | 0.218 (0.041-1.167) | 0.075 | 0.346 (0.046-2.572) | 0.299 |
| miR-196b | -- | - | 0.123 (0.020-.770) | 0.025 | 0.156 (0.034-0.715) | 0.017 |
| Age | 1.325 (0.458-3.837) | 0.604 | 0.891 (0.219-3.631) | 0.872 | 0.983 (0.205-4.709) | 0.983 |
| Gender | 0.517 (0.156-1.715) | 0.281 | 0.411 (0.091-1.863) | 0.249 | 0.912 (0.180-4.613) | 0.912 |
EFS: Event-free survival; DFS: Disease-free survival; OS: Overall survival; HR: Hazard ratio; CI: Confidence interval
Table 4 Multivariate analysis using miRNAs significantly affecting survival outcome in univariate analysis
Discussion
Pediatric AML is a heterogenous and complex disease in terms of cytogenetics and molecular landscapes. Involvement of miRNAs in various cancers, including pediatric AML has already been reported in many studies [14]. The focus of the current study was to identify dysregulated miRNA expression in pediatric CBF-AML patients, their interaction with predicted mRNA targets, and their potential significance as prognostic biomarkers. We found that the differentially expressed miRNAs can regulate the expression of a number of genes with potential involvement in AML pathogenesis via their involvement in multiple cellular processes like cell signalling, tumor suppression, epigenetics, transcription regulation, chromatin modifications, and myeloid transcription factor gene fusions or mutations. Our analysis demonstrates an upregulation of miR-100-5p, miR-4446-3p, and miR-335-3p while the expression of miR- 409-3p, miR-151a-3p, miR-196b, miR-4772-5p, and miR-758-3p was downregulated in pediatric CBF-AML patients as compared to controls. Although the miRNA expression profile in the present study is largely in line with our recent findings in pediatric CN-AML, certain differences were noted. While miR-196b and miR-1275 were upregulated in pediatric CN-AML patients [11], miR-196b was found to be downregulated in pediatric CBF-AML patients and miR-1275 levels remained unchanged. This suggests that while miRNA deregulation is a generalized phenomenon in pediatric AML, expression levels of certain miRNAs may be dependent on specific cytogenetic abnormalities.
As distinctly bimodal distribution or clustering of miR-758-3p and miR-4772-5p expression in pediatric CBF-AML patients was observed, their expression levels were also compared separately in these two distinct populations. The expression of miR-758-3p retained statistical significance (p<0.01) even when both populations were compared separately to pediatric controls. However, removing CBF-AML patient population with distinctly downregulated miR- 4772-5p expression from the analysis impacted the significance of miR-4772-5p expression in rest of the CBF-AML patients as compared to pediatric controls. Further, no significant differences in any clinical/molecular parameters as well as survival outcomes were observed between these distinct populations.
While miR-758-3p and miR-4446-3p had prognostic significance for pediatric CN-AML patients [11], we noted independent prognostic significance of miR-196b and miR-4772-5p for pediatric CBF-AML patients in the present study. There are very limited studies regarding the involvement of miR-4772-5p in other human malignancies. Though downregulation of serum exosomal miR-4772-3p has been suggested as a prognostic biomarker for tumor recurrence in stage II and stage III colon cancer patients [15], further studies are warranted to elucidate the role of miR-4772 in other human malignancies. Based on the evidence of downregulation of miR-196b and miR-4772-5p expression and their correlation with inferior survival outcome in CBF-AML patients, we speculate that they might have a tumor suppressor role. This is further supported by the fact that a common target of both these miRNAs, N-RAS is a known oncogene, and was found to be upregulated in CBF-AML patients.
A number of studies have reported dysregulated miRNA expression in AML and other human malignancies. The expression of miR-100-5p has been found to be dysregulated in various cancers, including AML [16–18]. Though increased expression of miR-100-5p has been associated with poor EFS and OS in pediatric AML [19], we did not observe any such correlation. This could be due to various factors such as relatively low sample size, different age groups, absence of significant number of patients with FLT3-ITD/TKD or NPM1 mutations, and different treatment modalities. While 67% of the 106 pediatric AML patients in the study by Bai et al., belonged to intermediate and/or adverse cytogenetic group, majority of the patients in our study exhibited favourable cytogenetics [19]. An upregulation of miR-335 expression in the present study is in line with earlier reports in adult AML and gastric cancer, but contrasts to it being downregulated in pediatric ALL [20-22].
We observed a downregulation of miR-196b-5p expression in our CBF-AML patient population, which is in contrast to other reports in epithelial ovarian cancer and lung cancer [23,24]. We found downregulation of miR-196b-5p to be associated with higher risk of relapse in pediatric CBF-AML patients. This is in contrast to an earlier report where upregulation of miR-196b-5p was associated with shorter OS in adult AML patients of intermediate-risk cytogenetics [25]. These differences could possibly be due to different age and cytogenetic risk groups of AML patients and different disease biology. Further, miR-196b-5p has also found to be overexpressed in myelodysplastic syndrome where it may be closely associated with the risk of transformation to leukemia [26].
Our report of downregulation of miR-409-3p expression in pediatric CBF-AML patients is consistent with other reports in prostate cancer, colorectal cancer, and bladder cancer, but in contrast to the reported upregulation of miR- 409-5p in t(15;17) pediatric AML [14,27-29]. In another study, lower miR-409-3p expression was associated with a higher risk of relapse in the intermediate-risk cytogenetic AML [25]. The expression of miR-151a-3p, which is downregulated in our study, has also been found to be downregulated in Imatinib-resistant chronic myeloid leukemia and ALL patients. In contrary, it was upregulated in cholangiocarcinoma and breast cancer and may inhibit metastasis in primary breast tumors [30,31]. We also observed downregulation of miR-758-3p in pediatric CBF-AML patients, which is in line with other studies in cervical cancer, papillary thyroid cancer and gastric cancer [32-34]. Similar to our data, upregulation of miR-4446-3p expression has been reported in breast cancer and cancer-associated fibroblast [35]. We did not find any deregulation of miR-589-5p, miR-1275, and miR-133b expression in our pediatric CBF-AML patients. However, a recent study concluded that miR-589-5p might act as a protective factor for AML patient survival [36]. Similarly, miR-1275 expression was downregulated in AML, while upregulation of miR-133b was observed in AML, lung cancer and esophageal squamous cell carcinoma [37-40]. In addition to the reasons stated earlier, the lack of homogeneity in the findings on the expression profile of individual miRNAs along with the prognostic significance they carry could be attributed to a number of factors including, but not restricted to i) differences in disease biology, ii) variability in the recruitment of number of patients and controls, iii) variability in the age and the frequency of distribution of patients in various cytogenetic and molecular risk groups, iv) variability in treatment modalities which can greatly impact the survival outcome, v) use of purified blast population for the downstream analysis, and vi) variability in the methods and analysis platform for miRNA profiling.
The prediction of genes targeted by individual miRNAs using bioinformatics approaches is more subjective and prone to variations among different studies, simply due to differences in the methods of analysis and more specifically due to the filters used for construction of the miRNA-mRNA networks. One of the major strengths of our study is that we not only predicted miRNA targets using in silico tools, but also analysed their expression in pediatric CBF-AML patients and controls. Our miRNA-mRNA interaction network (constructed based on expression profiles) revealed that a single miRNA can interact with multiple mRNAs, either directly or indirectly. For example, miR-4446-3p has been shown to downregulate tumor suppression-associated genes expression in breast cancer [41]. We identified and validated the expression of various predicted targets of miR-4446-3p, including PAX-5, BRAF, NRAS, KMT2A, RUNX1, ERBB2, IDH2, and CEBPA indicating its potential role in AML pathogenesis through regulating these targets. N-RAS and SF3B1 were predicted as potential targets of miR-4772-5p. SF3B1, which encodes for splicing factor 3B subunit 1, is located on 2q33.1 and is frequently altered in leukemia, suggesting its crucial role in disease initiation. Normal expression of BRAF might be due to the combined effects of upregulated miR-335-3p and downregulated miR-196b, and same is applicable for ERBB2, which is indirectly targeted by miR-4446-3p and miR-196b.
Conclusion
In conclusion, we analyzed the expression of 11 miRNAs, predicted targets of these miRNAs using in silico tools, studied their expression, and established a miRNA-mRNA network to find molecular pathways potentially involved in the pathogenesis of pediatric CBF-AML. We also identified important prognostic role of miR-4772-5p and miR- 196b. Along with a number of known miRNAs, our study reported deregulation in the expression of few less explored miRNAs such as miR-758-3p, miR-4446-3p, and miR-4772-5p, for which only limited data is available for their involvement in cancer, including AML. In conjunction with our earlier study in pediatric CN-AML, we hypothesize that the expression of at least some miRNAs may depend on the cytogenetic profile of pediatric AML patients. This needs to be validated using larger patient cohorts including well-defined cytogenetic presentations and their correlation with survival outcomes. It would be interesting to perform functional studies using in vitro and in vivo models to establish role of these miRNAs in AML pathogenesis.
Declerations
Author Contribution
SK, SB and PS designed the study. VG, SC and SA performed all the experiments and bioinformatics analysis. VG, SKS, PS and SK performed data analysis. SB and SC provided patient samples and clinical data. All authors were involved in manuscript writing, editing and final submission.
Funding/Acknowledgments
The study was supported by a research grant (Grant No. BT/PR8679/AGR/36/753/2013) from the Department of Biotechnology, Government of India, New Delhi, India. Vikas Gaur acknowledges Indian Council of Medical Research (ICMR), Government of India, New Delhi, India for the senior research fellowship (letter no. 3/2/59/2018/online Onco Fship/NCD III).
Conflict Of Interest
The authors declared no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
References
- Arber, Daniel A., et al. "The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia." Blood, Vol. 127, No. 20, 2016, pp. 2391-405.
- Döhner, Hartmut, et al. "Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel." Blood, Vol. 129, No. 4, 2017, pp. 424-47.
- Creutzig, Ursula, et al. "Changes in cytogenetics and molecular genetics in acute myeloid leukemia from childhood to adult age groups." Cancer, Vol. 122, No. 24, 2016, pp. 3821-30.
- Diederichs, Sven, and Daniel A. Haber. "Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression." Cell, Vol. 131, No. 6, 2007, pp. 1097-108.
- Lee, Yoontae, et al. "The nuclear RNase III Drosha initiates microRNA processing." Nature, Vol. 425, No. 6956, 2003, pp. 415-9.
- Lee, Yoontae, et al. "MicroRNA genes are transcribed by RNA polymerase II." The EMBO Journal, Vol. 23, No. 20, 2004, pp. 4051-60.
- Vaschetto, Luis M. "miRNA activation is an endogenous gene expression pathway." RNA Biology, Vol. 15, No. 6, 2018, pp. 826-8.
- Marcucci, Guido, et al. "MicroRNA expression profiling in acute myeloid and chronic lymphocytic leukaemias." Best Practice & Research Clinical Haematology, Vol. 22, No. 2, 2009, pp. 239-48.
- Bartel, David P. "MicroRNAs: genomics, biogenesis, mechanism, and function." Cell, Vol. 116, No. 2, 2004, pp. 281-97.
- Ambros, Victor. "The functions of animal microRNAs." Nature, Vol. 431, No. 7006, 2004, pp. 350-5.
- Gaur, Vikas, et al. "Dysregulation of miRNA expression and their prognostic significance in paediatric cytogenetically normal acute myeloid leukaemia." British Journal of Haematology, Vol. 188, No. 6, 2020, pp. e90-4.
- Chopra, Anita, et al. "Nucleophosmin mutation analysis in acute myeloid leukaemia: Immunohistochemistry as a surrogate for molecular techniques." The Indian Journal of Medical Research, Vol. 143, No. 6, 2016, pp. 763-8.
- Sharawat, Surender Kumar, et al. "High fms-like tyrosine kinase-3 (FLT3) receptor surface expression predicts poor outcome in FLT3 internal tandem duplication (ITD) negative patients in adult acute myeloid leukaemia: A prospective pilot study from India." The Indian Journal of Medical Research, Vol. 143, No. Suppl 1, 2016, pp. S11.
- Obulkasim, Askar, et al. "Classification of pediatric acute myeloid leukemia based on miRNA expression profiles." Oncotarget, Vol. 8, No. 20, 2017, pp. 33078-85.
- Liu, Chang, et al. "Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer." Oncotarget, Vol. 7, No. 46, 2016, pp. 76250-60.
- Guo, Peng, et al. "miR-100 resensitizes resistant epithelial ovarian cancer to cisplatin." Oncology Reports, Vol. 36, No. 6, 2016, pp. 3552-8.
- Shi, Wei, et al. "Significance of Plk1 regulation by miR‐100 in human nasopharyngeal cancer." International Journal of Cancer, Vol. 126, No. 9, 2010, pp. 2036-48.
- Liu, Jing, et al. "MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1." BMC Cancer, Vol. 12, No. 1, 2012, pp. 1-11.
- Bai, Jin, et al. "Upregulation of microRNA-100 predicts poor prognosis in patients with pediatric acute myeloid leukemia." OncoTargets and Therapy, Vol. 5, 2012, pp. 213-9.
- Yingchun, Li, et al. "Bone marrow MicroRNA-335 level predicts the chemotherapy response and prognosis of adult acute myeloid leukemia." Medicine, Vol. 94, No. 33, 2015.
- Li, Han, et al. "Long non-coding RNA MSTO2P promotes the proliferation and colony formation in gastric cancer by indirectly regulating miR-335 expression." Tumor Biology, Vol. 39, No. 6, 2017, pp. 1010428317705506.
- Yan, Junli, et al. "Deregulated MIR 335 that targets MAPK 1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia." British Journal of Haematology, Vol. 163, No. 1, 2013, pp. 93-103.
- Chong, Gun Oh, et al. "Overexpression of microRNA-196b accelerates invasiveness of cancer cells in recurrent epithelial ovarian cancer through regulation of homeobox A9." Cancer Genomics-Proteomics, Vol. 14, No. 2, 2017, pp. 137-41.
- Tellez, Carmen S., et al. "miR-196b is epigenetically silenced during the premalignant stage of lung carcinogenesis." Cancer Research, Vol. 76, No. 16, 2016, pp. 4741-51.
- Diaz-Beya, M., et al. "MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia." Leukemia, Vol. 28, No. 4, 2014, pp. 804-12.
- Wen, Jing, et al. "Over-expression of miR-196b-5p is significantly associated with the progression of myelodysplastic syndrome." International Journal of Hematology, Vol. 105, No. 6, 2017, pp. 777-83.
- Josson, Sajni, et al. "miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer." Clinical Cancer Research, Vol. 20, No. 17, 2014, pp. 4636-46.
- Liu, Mulin, et al. "Downregulation of microRNA-409-3p promotes aggressiveness and metastasis in colorectal cancer: an indication for personalized medicine." Journal of Translational Medicine, Vol. 13, No. 1, 2015, pp. 195.
- Xu, Xin, et al. "MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met." Molecules and Cells, Vol. 36, No. 1, 2013, pp. 62-8.
- McNally, Megan E., et al. "Concomitant dysregulation of microRNAs miR‐151‐3p and miR‐126 correlates with improved survival in resected cholangiocarcinoma." HPB, Vol. 15, No. 4, 2013, pp. 260-4.
- Krell, Jonathan, et al. "The clinico-pathologic role of microRNAs miR-9 and miR-151-5p in breast cancer metastasis." Molecular Diagnosis & Therapy, Vol. 16, No. 3, 2012, pp. 167-72.
- Meng, Xianhua, et al. "Regulatory roles of miRNA-758 and matrix extracellular phosphoglycoprotein in cervical cancer." Experimental and Therapeutic Medicine, Vol. 14, No. 4, 2017, pp. 2789-94.
- Chen, Jianping, et al. "MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1." Die Pharmazie-An International Journal of Pharmaceutical Sciences, Vol. 74, No. 4, 2019, pp. 235-8.
- Guo, Jinxing, et al. "Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer." Technology in Cancer Research & Treatment, Vol. 17, 2018, pp. 1533033818816061.
- Farina, Nicholas H., et al. "Development of a predictive miRNA signature for breast cancer risk among high-risk women." Oncotarget, Vol. 8, No. 68, 2017, pp. 112170.
- Zhang, Chunmei, et al. "Identification of miRNA-mRNA network associated with acute myeloid leukemia survival." Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, Vol. 23, 2017, pp. 4705.
- Ozdogan, Hakan, et al. "DICER1 gene and miRNA dysregulation in mesenchymal stem cells of patients with myelodysplastic syndrome and acute myeloblastic leukemia." Leukemia Research, Vol. 63, 2017, pp. 62-71.
- Niederwieser, Christian, et al. "Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia." Leukemia, Vol. 29, No. 3, 2015, pp. 567-75.
- Mi, Yonghua, Miao He, and Beizhong Liu. "MiR-133b Affect the Proliferation and Drug Sensitivity in A549 Lung Cancer Stem Cells by Targeting PKM2." Zhongguo fei ai za zhi= Chinese Journal of Lung Cancer, Vol. 20, No. 6, 2017, pp. 376-81.
- Qin, Yi, et al. "SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma." Acta Biochimica et Biophysica Sinica, Vol. 49, No. 2, 2017, pp. 138-48.
- Kim, Baek Gil, et al. "Transcriptome-wide analysis of compression-induced microRNA expression alteration in breast cancer for mining therapeutic targets." Oncotarget, Vol. 7, No. 19, 2016, pp. 27468.