Research - International Journal of Medical Research & Health Sciences ( 2022) Volume 11, Issue 5
ACE I/D Gene Polymorphism and Short-Term Endurance Training Response of Untrained Individuals
Hazwani Ahmad Yusof*Hazwani Ahmad Yusof, Institute of Advanced Medicine and Dentistry, Lifestyle Science Cluster, Advanced Medical and Dental Institute, Universiti Sains Malaysia, Kepala Batas, Malaysia, Email: hazwanihanafi@usm.my
Received: 17-May-2022, Manuscript No. ijmrhs-22-64042 ; Editor assigned: 19-May-2022, Pre QC No. ijmrhs-22-64042 (PQ); Reviewed: 24-May-2022, QC No. ijmrhs-22-64042 (Q); Revised: 24-May-2022, Manuscript No. ijmrhs-22-64042 (R); Published: 31-May-2022
Abstract
We investigate whether ACE I/D gene polymorphism modulates anthropometric, cardiovascular, biochemical, OGTT, and fat oxidation responses to six weeks of short-term endurance training for untrained adults. Thirty-five untrained healthy individuals initially participated in the pre-test assessment and were screened for ACE I/D genotype. However, only seventeen participants completed three sessions of 45 minutes of endurance training every week for 4 weeks. Using two-way repeated measure ANOVA, the data were clustered among the three classes of ACE I/D genotype groups (II=3, ID=7, DD=7). Six weeks of endurance training resulted in increased the VO2 max (p=0.004), fat oxidation at 40% (p=0.028), 50% (p<0.001), 60% (p=0.011) and 70% (p<0.001) of VO2 max. A significant interaction effect between ACE and endurance training on the fat oxidation variables has been reported (F(18, 12)=2.701, p=0.042; Wilks’ Λ=0.039). The interaction observed showed that while the ID genotype group had a much higher fat oxidation score before endurance training, the II genotype group had the highest score after the training program. The present study found that short-term endurance training may be favorable for certain ACE I/D genotypes.
Keywords
ACE I/D gene polymorphism, Endurance, Fat oxidation, Cardiovascular, Short-term, Untrained
Introduction
Previous studies on the association of ACE I/D gene polymorphism and response to endurance training have led to several findings [1-6]. While some studies have shown a connection between the presence of the I allele with enhancement in endurance performance following the endurance training, similar results could not be replicated by other studies [1,3,6-10]. The choice of samples that may have a prolonged training impact, such as athletes and army recruits in those studies reporting a lack of association, may underlie this contradictory result. Additional studies among untrained participants are therefore required to evaluate the effect of ACE I/D gene polymorphism on improved training outcomes to minimize gene-environmental interactions that could modify training responses.
Despite an association with endurance performance, polymorphism of the ACE I/D gene was also reported to be correlated with glucose tolerance [11-17]. One of the genetic risks of diabetic nephropathy and glucose intolerance is indicated by ACE I/D gene polymorphism due to elevated plasma ACE activity in certain diabetic patients [8,13,18]. In the oral glucose tolerance test, participants with the D allele have been found to have higher blood glucose levels than those with I allele carriers [12]. Concurrent with positive findings from several studies that have shown that exercise training improves tolerance to glucose, particularly among patients with diabetes, some studies investigated whether ACE I/D gene polymorphism can modulate the effect of glucose tolerance exercise training [11,14-17,19,20]. Dengel, et al. evaluated the association between insulin resistance and the ACE I/D gene polymorphism in a group of older hypertensive participants before and after 6 months of the Aerobic Exercise Program (AEX) [19]. Dengel, et al. reported that there was a significant interaction between AEX and ACE I/D genotype, where glucose-insulin reactions were significantly lower among II genotype carriers than those of DD and ID genotypes [19]. In addition, Hurlbut, et al. investigated whether the effects of the 6 months of full-body strength training on the glucose homeostasis were affected by the age, sex, or ACE I/D genotype and found that those with at least one I allele appeared to decrease the total insulin than D allele [20]. While these two studies used a prolonged intervention program, the association of ACE I/D gene polymorphism with glucose response to short-term endurance training was not well known. The association knowledge can help to determine who is most likely to boost glucose tolerance with short-term endurance training and can also support a personalized diabetic management training program.
The ACE I/D gene polymorphism has also been related to anthropometric, cardiovascular, and biochemical parameters for the development of cardiovascular and metabolic diseases [9,21]. DD genotype frequency and D allele frequency of the 1762 Hungarian population were found to be significantly higher in the metabolic group than in the nonmetabolic group [9]. Compared to other ACE I/D genotypes, the II genotype had lower serum total cholesterol levels, low-density lipoprotein cholesterol levels, and non-high-density lipoprotein cholesterol levels in a study group of 341 men aged 39 to 65 years, indicating that the I allele has a protective effect against high serum lipid and lipoprotein levels [a href="#21" title="21">21]. ACE I/D gene polymorphism has been found by Hagberg, et al. to be associated with variable responses in the clinical population via prolonged endurance training [10]. Although Hagberg, et al. have been engaged through extended endurance training, it is not clear whether the short-term endurance training impact on anthropometric and lipid reactions would also be modulated by ACE I/D gene polymorphism [10].
The present study was therefore undertaken to determine whether the ACE I/D gene polymorphism correlated with anthropometric, cardiovascular, biochemical, OGTT, and fat oxidation responses to 4 weeks of short-term endurance training of untrained individuals. We assumed that after 4 weeks of short-term endurance training, anthropometric, cardiovascular, biochemical, OGTT, and fat oxidation reactions in certain ACE I/D genotype groups were thought to have improved, especially among I allele carriers, compared to D allele carriers.
Materials and Methods
Study Design
A repeated measures design was used in the present study. The following parameters were measured before and after 4 weeks of short-term endurance training: anthropometric, cardiovascular, biochemical, blood glucose during oral glucose tolerance test, and fat oxidation, The study protocol is accepted by the Human Research Ethics Committee of the University of Sydney, Australia, in compliance with the ethical principles of the Helsinki Declaration.
Participants
The sample size was calculated using the Power and Sample Size (P.S.) software. The power of the study is set at 0.80 and the type I error probability associated with this test of this null hypothesis is 0.05. A total of 35 healthy male and female untrained individuals capable to perform a cycling test to exhaustion have initially participated in the pretesting evaluation. However, due to individual reasons, only seventeen participants (4 females, 13 males), 22.7 ± 2.8 years old, from the initial sample of 35 had participated in the short-term endurance training program and completed the post-training evaluation.
Preliminary Assessments
After the participants were briefed on the research procedures, a written consent form was obtained from all participants. Participants were asked to provide a 5 ml blood sample to evaluate the ACE I/D genotype and to familiarise them with the test protocols.
ACE I/D Genotyping Determination
Genomic DNA was isolated from the blood samples using the Wizard® Genomic DNA Purification Kit following the manufacturer’s protocol (Promega Corporation, United States of America). Polymerase Chain Reaction (PCR) was carried out in a final volume of 25 μl consisting of 2.5 μl of 10X standard reaction buffer (GeneAllBiotechnologyCo. Ltd, Korea) (25 mM Mg2+, 50 mM Tris-HCl, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 50% glycerol), 2.0 μl of dNTP mix (200 μM each dNTP (dATP, dCTP, dGTP, dDTP)), 0.8 μM of each primer (forward primer: 5’-CTGGAGACCACTCCCATCCTTTCT-3’: reverse primer: 5’-CTGGAGACCACTCCCATCCTTTCT-3’), 0.5 units of Taq DNA polymerase, 2.5 μl of dimethylsulfoxide, 10.8 μl of sterilize distilled water, and 5 μl of genomic DNA. The target fragment bearing the ACE I/D polymorphism was amplified under the following conditions: 7 minutes at 95°C followed by 25 cycles of 30 seconds at 95°C, 30 seconds at 62°C, and 1 minute at 72°C, with a final step of 7 minutes at 72°C. The amplified products were electrophoresed on a 1.5% agarose gel that was pre-stained with ethidium bromide at 70 volts for 1 hour. The presence of 490 and 190 base pair bands indicated the ACE insertion (I) and deletion (D) alleles, respectively.
Pre- and Post-Training Tests
A total of 35 participants participated in the pre-testing evaluation whilst 17 participants from the initial sample of 35 completed the post-training evaluation. Participants reported to the laboratory in the morning after an overnight fast. Bodyweight, height, body mass index, waist and hip circumference, heart rate, blood pressure, cholesterol, highdensity lipoprotein, and low-density lipoprotein were measured before the oral glucose tolerance test. The participants consumed 75 g of glucose in a total volume of 200 ml of water within 2 minutes. A 1.0 ml blood sample was taken via a finger prick at 7-time points following the glucose load (0, 10, 20, 30, 60, 90, and 120) min. Following the Oral Glucose Tolerance Test (OGTT), the participants performed maximal oxygen consumption (VO2 max) testing on a cycle ergometer to measure their VO2 max and fat oxidation rate at (20%, 30%, 40%, 50%, 60%, and 70%) VO2 max. Before the VO2 max test, the participants pedal on the cycle ergometer at three different sub-maximal exercise workloads with each stage lasting for 7 minutes then followed by 5 minutes of very low-intensity recovery cycling. Starting with 100 watts, the ergometer power was increased by 15 watts every 30 seconds until the participants were unable to continue the cycling exercise despite verbal encouragement from the researcher.
Endurance Training
The participants were trained on a cycle ergometer, and the training was tailored to each participant’s aerobic capacity using the data collected from the VO2 max test performed at the pre-training test. The endurance training consisted of 45 minutes of cycling per session 3 times per week for 4 weeks. In the first two weeks, the participants cycled at intensities corresponding to 60% of their VO2 max, whereas in the third week, they cycled at 70% VO2 max and 75% of VO2 max in the fourth week of the intervention. The training protocol used in this study was based on previously published protocols by Reardon, et al. [22].
Statistical Analysis
All statistical evaluations were performed using the IBM SPSS statistical version 20.0, United States, with the level of significance set at p<0.05. The descriptive information is presented as mean ± SD. The studied variables at pre- and post-measurement were analyzed with one-way ANOVA, and then with the LSD or Games-Howell correction post hoc test, of comparing all the parameters between ACE I/D genotype groups over the training periods where appropriate. The analysis of the changes in parameters before and after training was carried out with a paired t-test. Positive and negative outcomes showed a rise and decline in endurance training, respectively. Two-way repeated ANOVA analysis measurements were used to determine the interaction of the ACE I/D genotype with the performance of the variables studied (Interaction: genotype x training; training effect (subject-in-subject) and genotype effect (entre-subject). The main effects of training and genotype are presented as estimated marginal mean ± standard error of the mean (SEM).
Results
Baseline Data
Thirty-five participants were originally recruited for and performed a pre-test. Of the thirty-five participants, 22 participants were male, and 13 participants were female. The characteristic of the overall participants obtained before the test is shown in Table 1. The OGTT value of all participants obtained before the test was within normal limits according to the WHO criteria indicating normal glucose tolerance in overall participants [23].
| Variables | Mean ± SD |
|---|---|
| Age (year) | 22.69 ± 3.43 |
| Anthropometric | |
| Height (cm) | 172.88 ± 9.96 |
| Body Weight (kg) | 70.68 ± 10.65 |
| Body Mass Index (kg/m2) | 23.58 ± 2.46 |
| Waist (cm) | 77.84 ± 6.50 |
| Hip Circumference (cm) | 97.25 ± 5.62 |
| Waist Hip Ratio | 0.80 ± 0.04 |
| Body Fat (%) | 20.73 ± 7.37 |
| Cardiovascular | |
| Systolic Blood Pressure (mmHg) | 115.09 ± 9.56 |
| Diastolic Blood Pressure (mmHg) | 71.80 ± 7.21 |
| Mean Arterial Pressure (mmHg) | 86.23 ± 6.62 |
| Resting Heart Rate (bpm) | 54.58 ± 9.93 |
| Resting Pulse Pressure (mmHg) | 6276.80 ± 1200.36 |
| Maximal Oxygen Consumption (L/min) | 3.05 ± 0.66 |
| Peak Power Output (Watts) | 259.66 ± 64.35 |
| Biochemical | |
| Triglyceride (mmol/L) | 0.80 ± 0.18 |
| Total Cholesterol (mmol/L) | 3.95 ± 0.63 |
| High Density Lipoprotein (mmol/L) | 1.35 ± 0.34 |
| Low Density Lipoprotein (mmol/L) | 2.06 ± 0.50 |
| Oral Glucose Tolerance Test | |
| Fasting Blood Glucose (mmol/L) | 4.96 ± 0.43 |
| Blood Glucose 30 minutes (mmol/L) | 8.53 ± 1.61 |
| Blood Glucose 60 minutes (mmol/L) | 7.40 ± 1.56 |
| Blood Glucose 90 minutes (mmol/L) | 6.82 ± 1.12 |
| Blood Glucose 120 minutes (mmol/L) | 5.97 ± 1.04 |
| Area Under Curve (mmol/L.min) | 14.12 ± 1.86 |
| Fat Oxidation | |
| Resting Fat Oxidation (gmin-1) | 0.07 ± 0.03 |
| Fat Oxidation at 20% VO2max (gmin-1) | 0.23 ± 0.07 |
| Fat Oxidation at 30% VO2max (gmin-1) | 0.23 ± 0.07 |
| Fat Oxidation at 40% VO2max (gmin-1) | 0.23 ± 0.09 |
| Fat Oxidation at 50% VO2max (gmin-1) | 0.20 ± 0.10 |
| Fat Oxidation at 60% VO2max (gmin-1) | 0.17 ± 0.12 |
| Fat Oxidation at 70% VO2max (gmin-1) | 0.12 ± 0.12 |
For the ACE I/D gene polymorphism, the allele frequencies in the overall group were 0.51 and 0.49 for the I and D alleles, respectively. The frequency of II, ID and DD genotypes in the overall participants were 25.7% (n=9), 51.4% (n=18) and 22.9% (n=8), respectively. The ACE I/D gene polymorphism frequency distribution was in Hardy Weinberg equilibrium (p>0.05)
The baseline characteristic data of the overall participants according to ACE I/D genotype are shown in Table 2. There were no significant differences between ACE I/D genotype groups in all variables (p>0.05), except HDL (F(2, 34)=3.658, p=0.037). Post hoc tests using the LSD correction found that the HDL was significantly higher in the ID genotype group (1.49 ± 0.33) when compared to the DD genotype group (1.16 ± 0.35) (p=0.021), though it was not significantly different from the values among II genotype group (1.24 ± 0.27) (p=0.066).
| Variables | II (n=9) | ID (n=18) | DD (n=8) | p-value |
|---|---|---|---|---|
| Age (year) | 22.89 ± 4.51 | 22.83 ± 3.43 | 22.13 ± 2.17 | 0.876 |
| Anthropometric | ||||
| Height (cm) | 172.99 ± 9.81 | 171.78 ± 11.29 | 175.23 ± 7.22 | 0.729 |
| Body Weight (kg) | 68.07 ± 10.35 | 70.22 ± 10.08 | 74.64 ± 12.43 | 0.444 |
| Body Mass Index (kg/m2) | 22.73 ± 2.77 | 23.72 ± 1.87 | 24.23 ± 3.26 | 0.443 |
| Waist (cm) | 77.23 ± 5.48 | 77.31 ± 6.24 | 79.73 ± 8.43 | 0.660 |
| Hip Circumference (cm) | 94.91 ± 4.58 | 98.13 ± 5.57 | 97.89 ± 6.69 | 0.361 |
| Waist Hip Ratio | 0.81 ± 0.04 | 0.79 ± 0.05 | 0.81 ± 0.04 | 0.246 |
| Body Fat (%) | 18.81 ± 5.60 | 22.22 ± 7.84 | 19.54 ± 8.17 | 0.472 |
| Cardiovascular | ||||
| Systolic Blood Pressure (mmHg) | 113.44 ± 9.93 | 114.22 ± 10.45 | 118.88 ± 6.66 | 0.477 |
| Diastolic Blood Pressure (mmHg) | 73.00 ± 6.78 | 69.67 ± 6.27 | 75.25 ± 8.78 | 0.162 |
| Mean Arterial Pressure (mmHg) | 86.48 ± 7.08 | 84.52 ± 5.92 | 89.79 ± 6.97 | 0.173 |
| Resting Heart Rate (beat per minute) | 54.78 ± 12.86 | 55.08 ± 9.44 | 53.23 ± 8.40 | 0.912 |
| Resting Pulse Pressure (mmHg) | 6142.46 ± 1130.56 | 6316.83 ± 1315.98 | 6337.88 ± 1140.32 | 0.930 |
| Maximal Oxygen Consumption (L/min) | 2.98 ± 0.68 | 3.01 ± 0.70 | 3.23 ± 0.59 | 0.686 |
| Peak Power Output (Watts) | 267.56 ± 79.74 | 253.83 ± 65.18 | 263.88 ± 48.41 | 0.861 |
| Biochemical | ||||
| Triglyceride (mmol/L) | 0.82 ± 0.18 | 0.74 ± 0.13 | 0.90 ± 0.24 | 0.108 |
| Total Cholesterol (mmol/L) | 3.62 ± 0.60 | 4.15 ± 0.51 | 3.85 ± 0.79 | 0.096 |
| High Density Lipoprotein (mmol/L) | 1.24 ± 0.27 | 1.49 ± 0.33 | 1.16 ± 0.35 | 0.037* |
| Low Density Lipoprotein (mmol/L) | 1.90 ± 0.39 | 2.09 ± 0.50 | 2.19 ± 0.61 | 0.470 |
| Oral Glucose Tolerance Test | ||||
| Fasting Blood Glucose (mmol/L) | 5.07 ± 0.56 | 4.93 ± 0.42 | 4.93 ± 0.27 | 0.712 |
| Blood Glucose 30 minutes (mmol/L) | 8.78 ± 1.67 | 8.27 ± 1.56 | 8.86 ± 1.77 | 0.611 |
| Blood Glucose 60 minutes (mmol/L) | 8.43 ± 2.22 | 6.99 ± 1.13 | 7.15 ± 1.09 | 0.063 |
| Blood Glucose 90 minutes (mmol/L) | 7.56 ± 1.47 | 6.57 ± 1.00 | 6.58 ± 0.48 | 0.069 |
| Blood Glucose 120 minutes (mmol/L) | 6.21 ± 1.40 | 5.87 ± 0.86 | 5.93 ± 1.04 | 0.723 |
| Area Under Curve (mmol/L.min) | 15.20 ± 2.63 | 13.62 ± 1.55 | 14.01 ± 0.98 | 0.111 |
| Fat Oxidation | ||||
| Resting Fat Oxidation (gmin-1) | 0.07 ± 0.04 | 0.06 ± 0.03 | 0.08 ± 0.03 | 0.552 |
| Fat Oxidation at 20% VO2max (gmin-1) | 0.22 ± 0.06 | 0.22 ± 0.08 | 0.24 ± 0.04 | 0.776 |
| Fat Oxidation at 30% VO2max (gmin-1) | 0.21 ± 0.05 | 0.23 ± 0.07 | 0.25 ± 0.08 | 0.392 |
| Fat Oxidation at 40% VO2max (gmin-1) | 0.20 ± 0.07 | 0.24 ± 0.08 | 0.24 ± 0.12 | 0.538 |
| Fat Oxidation at 50% VO2max (gmin-1) | 0.17 ± 0.09 | 0.21 ± 0.10 | 0.22 ± 0.10 | 0.491 |
| Fat Oxidation at 60% VO2max (gmin-1) | 0.09 ± 0.12 | 0.14 ± 0.13 | 0.11 ± 0.11 | 0.231 |
| Fat Oxidation at 70% VO2max (gmin-1) | 0.09 ± 0.12 | 0.14 ± 0.13 | 0.11 ± 0.11 | 0.594 |
| Data are shown as mean ± SD; * p<0.05; Based on LSD comparison, ID vs. DD | ||||
The current research included both female and male participants. As a result, data were analyzed by gender to monitor the possible impact of gender in the data analysis. The data for each gender category is provided in Table 3. In the female group, there were no significant differences between ACE I/D genotype groups in all anthropometric, cardiovascular, and biochemical variables (p>0.05), except for waist-hip ratio (F(2, 12)=9.532, p=0.005). Post hoc tests using the Games-Howell correction revealed that the waist-hip ratio was significantly lower in the ID genotype group (0.75 ± 0.02) when compared to the DD genotype group (0.84 ± 0.06) (p=0.002), though it was not significantly different with the values among II genotype group (0.80 ± 0.00) (p=0.59).
| Variables | Female (n = 13) | Male (n = 22) | ||||||
|---|---|---|---|---|---|---|---|---|
| II (n=2) | ID (n=9) | DD (n=2) | p-value | II (n=7) | ID (n=9) | DD (n=6) | p-value | |
| Age (year) | 22.00 ± 2.83 | 20.78 ± 2.33 | 24.00 ± 1.41 | 0.24 | 23.14 ± 5.05 | 24.89 ± 3.18 | 21.50 ± 2.07 | 0.235 |
| Anthropometric | ||||||||
| Height (cm) | 164.55 ± 11.10 | 163.50 ± 7.14 | 171.45 ± 11.67 | 0.486 | 175.40 ± 8.79 | 180.06 ± 8.11 | 176.48 ± 6.18 | 0.477 |
| Body Weight (kg) | 62.55 ± 6.86 | 63.10 ± 6.62 | 74.10 ± 28.85 | 0.456 | 69.64 ± 11.05 | 77.34 ± 7.63 | 74.82 ± 7.04 | 0.238 |
| Body Mass Index (kg/m2) | 23.09 ± 0.58 | 23.56 ± 1.55 | 24.71 ± 6.44 | 0.789 | 22.63 ± 3.18 | 23.88 ± 2.24 | 24.07 ± 2.54 | 0.555 |
| Waist (cm) | 75.50 ± 6.36 | 74.10 ± 4.17 | 84.10 ± 18.95 | 0.265 | 77.73 ± 5.66 | 80.52 ± 6.49 | 78.27 ± 4.17 | 0.588 |
| Hip Circumference (cm) | 94.95 ± 8.13 | 98.88 ± 5.92 | 99.80 ± 15.56 | 0.781 | 94.90 ± 4.11 | 97.38 ± 5.45 | 97.25 ± 3.51 | 0.525 |
| Waist Hip Ratio | 0.80 ± 0.00 | 0.75 ± 0.02 | 0.84 ± 0.06 | 0.005* | 0.82 ± 0.04 | 0.83 ± 0.03 | 0.80 ± 0.03 | 0.531 |
| Body Fat (%) | 26.93 ± 2.75 | 28.01 ± 4.13 | 29.51 ± 8.22 | 0.854 | 16.49 ± 3.51 | 16.43 ± 6.19 | 16.21 ± 5.18 | 0.995 |
| Cardiovascular | ||||||||
| Systolic Blood Pressure (mmHg) | 99.00 ± 1.41 | 110.67 ± 9.37 | 110.00 ± 0.00 | 0.248 | 117.57 ± 6.45 | 117.78 ± 10.77 | 121.83 ± 4.49 | 0.581 |
| Diastolic Blood Pressure (mmHg) | 65.50 ± 7.78 | 69.78 ± 6.98 | 66.00 ± 8.49 | 0.66 | 75.14 ± 5.21 | 69.56 ± 5.90 | 78.33 ± 6.92 | 0.032* |
| Mean Arterial Pressure (mmHg) | 76.67 ± 4.71 | 83.41 ± 6.06 | 80.67 ± 5.66 | 0.463 | 89.29 ± 4.66 | 85.63 ± 5.92 | 92.83 ± 4.15 | 0.046* |
| Resting Heart Rate (beat per minute) | 68.00 ± 18.38 | 58.99 ± 9.65 | 53.75 ± 8.13 | 0.427 | 51.00 ± 9.45 | 51.17 ± 7.87 | 53.06 ± 9.25 | 0.896 |
| Resting Pulse Pressure (mmHg) | 6745.00 ± 1916.26 | 6570.01 ± 1394.85 | 5912.50 ± 894.49 | 0.812 | 5970.30 ± 967.79 | 6063.64 ± 1261.05 | 6479.67 ± 1250.58 | 0.714 |
| Maximal Oxygen Consumption (L/min) | 2.56 ± 0.42 | 2.44 ± 0.47 | 2.86 ± 1.03 | 0.632 | 3.11 ± 0.71 | 3.58 ± 0.29 | 3.36 ± 0.46 | 0.201 |
| Peak Power Output (Watts) | 199.00 ± 32.53 | 204.22 ± 42.36 | 223.50 ± 79.90 | 0.847 | 287.14 ± 79.29 | 303.44 ± 41.17 | 277.33 ± 33.69 | 0.653 |
| Biochemical | ||||||||
| Triglyceride (mmol/L) | 0.69 ± 0.15 | 0.71 ± 0.12 | 1.00 ± 0.25 | 0.327 | 0.86 ± 0.18 | 0.78 ± 0.14 | 0.87 ± 0.25 | 0.585 |
| Total Cholesterol (mmol/L) | 4.46 ± 0.04 | 4.20 ± 0.50 | 4.80 ± 0.12 | 0.267 | 3.38 ± 0.42 | 4.11 ± 0.53 | 3.53 ± 0.63 | 0.029* |
| High Density Lipoprotein (mmol/L) | 1.57 ± 0.03 | 1.63 ± 0.35 | 1.54 ± 0.45 | 0.925 | 1.15 ± 0.22 | 1.35 ± 0.25 | 1.04 ± 0.23 | 0.058 |
| Low Density Lipoprotein (mmol/L) | 2.19 ± 0.12 | 1.94 ± 0.50 | 2.80 ± 0.45 | 0.107 | 1.81 ± 0.41 | 2.24 ± 0.49 | 1.99 ± 0.53 | 0.218 |
| Oral Glucose Tolerance Test | ||||||||
| Fasting Blood Glucose (mmol/L) | 5.60 ± 0.14 | 4.94 ± 0.58 | 5.05 ± 0.35 | 0.327 | 4.91 ± 0.54 | 4.91 ± 0.22 | 4.88 ± 0.26 | 0.986 |
| Blood Glucose 30 minutes (mmol/L) | 10.25 ± 1.48 | 7.99 ± 1.20 | 8.90 ± 3.11 | 0.203 | 8.36 ± 1.56 | 8.54 ± 1.89 | 9.85 ± 1.56 | 0.874 |
| Blood Glucose 60 minutes (mmol/L) | 9.25 ± 1.20 | 7.29 ± 0.85 | 7.85 ± 0.64 | 0.048* | 8.20 ± 2.46 | 6.70 ± 1.34 | 6.92 ± 1.14 | 0.225 |
| Blood Glucose 90 minutes (mmol/L) | 7.70 ± 1.70 | 6.74 ± 1.03 | 6.15 ± 0.21 | 0.37 | 7.51 ± 1.55 | 6.39 ± 0.99 | 6.72 ± 0.46 | 0.153 |
| Blood Glucose 120 minutes (mmol/L) | 7.00 ± 0.85 | 6.20 ± 0.81 | 5.40 ± 0.42 | 0.176 | 5.99 ± 1.49 | 5.53 ± 0.81 | 6.10 ± 1.16 | 0.599 |
| Area Under Curve (mmol/L.min) | 16.75 ± 2.44 | 13.82 ± 1.22 | 14.06 ± 1.96 | 0.081 | 14.76 ± 2.68 | 13.43 ± 1.87 | 13.99 ± 0.77 | 0.425 |
| Fat Oxidation | ||||||||
| Resting Fat Oxidation (gmin-1) | 0.02 ± 0.01 | 0.06 ± 0.02 | 0.05 ± 0.01 | 0.07 | 0.08 ± 0.04 | 0.06 ± 0.04 | 0.09 ± 0.02 | 0.414 |
| Fat Oxidation at 20% VO2max (gmin-1) | 0.19 ± 0.06 | 0.20 ± 0.03 | 0.26 ± 0.09 | 0.222 | 0.23 ± 0.06 | 0.25 ± 0.11 | 0.24 ± 0.02 | 0.938 |
| Fat Oxidation at 30% VO2max (gmin-1) | 0.18 ± 0.04 | 0.22 ± 0.06 | 0.29 ± 0.10 | 0.242 | 0.22 ± 0.05 | 0.25 ± 0.08 | 0.24 ± 0.08 | 0.652 |
| Fat Oxidation at 40% VO2max (gmin-1) | 0.16 ± 0.01 | 0.21 ± 0.08 | 0.29 ± 0.10 | 0.285 | 0.21 ± 0.07 | 0.26 ± 0.08 | 0.22 ± 0.13 | 0.944 |
| Fat Oxidation at 50% VO2max (gmin-1) | 0.12 ± 0.01 | 0.23 ± 0.12 | 0.29 ± 0.09 | 0.335 | 0.18 ± 0.10 | 0.20 ± 0.09 | 0.20 ± 0.11 | 0.648 |
| Fat Oxidation at 60% VO2max (gmin-1) | 0.09 ± 0.01 | 0.23 ± 0.13 | 0.26 ± 0.07 | 0.28 | 0.14 ± 0.10 | 0.17 ± 0.09 | 0.12 ± 0.14 | 0.648 |
| Fat Oxidation at 70% VO2max (gmin-1) | 0.06 ± 0.02 | 0.19 ± 0.15 | 0.24 ± 0.06 | 0.391 | 0.10 ± 0.14 | 0.09 ± 0.08 | 0.07 ± 0.08 | 0.882 |
| Data are shown as mean ± SD; * p<0.05 | ||||||||
Meanwhile, in the male group, there were no significant differences between ACE I/D genotype groups in all variables (p>0.05), except for diastolic blood pressure (F(2, 21)=4.160, p=0.032), mean arterial pressure (F(2, 21)=3.624, p=0.046), and total cholesterol (F(2, 21)=4.308, p=0.029). Post hoc tests using the LSD correction found that the diastolic blood pressure was significantly lower in the ID genotype group (69.56 ± 5.90) when compared to the DD genotype group (78.33 ± 6.92) (p=0.012), though it was not significantly different with the values among II genotype group (75.14 ± 5.21) (p=0.08). For mean arterial pressure, post hoc tests using the LSD correction revealed that the mean arterial pressure was significantly lower in the ID genotype group (85.63 ± 5.92) when compared to the DD genotype group (92.83 ± 4.15) (p=0.015), though it was not significantly different with the values among II genotype group (89.29 ± 4.66) (p=0.172). Meanwhile, for total cholesterol, post hoc tests using the Games-Howell correction revealed that the mean total cholesterol was significantly lower in the II genotype group (3.38 ± 0.42) when compared to the ID genotype group (4.11 ± 0.53) (p=0.021) though it was not significantly different with the values DD genotype group (3.53 ± 0.63) (p=0.871).
There were no significant differences between ACE I/D genotype groups in all OGTT values (p<0.05) among the male group. In the female group, there were no significant differences between ACE I/D genotype groups in all variables (p>0.05), except for blood glucose value at 60 minutes (F(2, 12)=4.177, p=0.048). Post hoc tests using the LSD correction found that blood glucose value at 60 minutes was significantly higher in the II genotype group (9.25 ± 1.20) relative to the ID genotype group (7.29 ± 0.85) (p=0.017), although it was not significantly different with the values among DD genotype group (7.85 ± 0.64) (p=0.14). There were no significant differences between ACE I/D genotype groups in all fat oxidation values in both female and male groups (p>0.05).
Responses to Exercise Training
Of the thirty-five initial participants, only seventeen participants (4 females, 13 males), aged 22.65 ± 2.78 years old had participated in the short-term endurance training program and completed the post-training evaluation. Table 4 shows the dependent variable responses in the whole sample (regardless of their ACE I/D genotype) following 4 weeks of endurance exercise training. The paired t-test revealed no significant effect of 4 weeks of endurance training on all dependent variables (p>0.05) except for VO2 max, fat oxidation at 40%, 50%, 60%, and 70% VO2 max as well as peak power output. VO2 max (t(16)=3.398, p=0.004), fat oxidation at 40% (t(16)=2.423, p=0.028), 50% (t(16)=4.446, p<0.001), 60% (t(16)=2.891, p=0.011) and 70% (t(16)=6.005, p<0.001) VO2 max were significantly increased following the six weeks of endurance training.
| Variables | Pre-Training | Post-Training | Change (Δ) with training | t-value | p-value |
|---|---|---|---|---|---|
| Anthropometric | |||||
| Body Weight (kg) | 73.65 ± 12.04 | 71.69 ± 14.72 | -1.97 ± 7.97 | -1.018 | 0.324 |
| Body Mass Index (kg/m2) | 23.88 ± 2.82 | 23.22 ± 3.79 | -0.66 ± 2.54 | -1.070 | 0.301 |
| Waist (cm) | 79.75 ± 7.94 | 79.14 ± 9.13 | -0.62 ± 2.69 | -0.946 | 0.358 |
| Hip Circumference (cm) | 98.02 ± 5.72 | 96.89 ± 6.54 | -1.13 ± 3.01 | -1.548 | 0.141 |
| Waist Hip Ratio | 0.81 ± 0.04 | 0.82 ± 0.05 | -0.00 ± 0.03 | 0.434 | 0.670 |
| Body Fat (%) | 19.24 ± 7.21 | 19.57 ± 7.61 | +0.33 ± 2.61 | 0.523 | 0.608 |
| Cardiovascular | |||||
| Systolic Blood Pressure (mmHg) | 120.06 ± 6.31 | 118.71 ± 6.02 | -1.35 ± 5.22 | -1.069 | 0.301 |
| Diastolic Blood Pressure (mmHg) | 74.18 ± 7.16 | 76.06 ± 6.70 | 1.88 ± 8.81 | 0.881 | 0.391 |
| Mean Arterial Pressure (mmHg) | 89.47 ± 5.69 | 90.27 ± 5.38 | 0.80 ± 6.25 | 0.530 | 0.603 |
| Resting Heart Rate (beat per minute) | 55.08 ± 7.49 | 53.17 ± 8.40 | -1.91 ± 6.04 | -1.304 | 0.211 |
| Resting Pulse Pressure (mmHg) | 6605.87 ± 924.93 | 6306.84 ± 1006.40 | -299.03 ± 807.07 | -1.528 | 0.146 |
| Maximal Oxygen Consumption (L/min) | 3.17 ± 0.67 | 3.38 ± 0.72 | 0.20 ± 0.25 | 3.398 | 0.004* |
| Peak Power Output (Watts) | 267.53 ± 61.16 | 292.71 ± 61.01 | 25.18 ± 17.29 | 6.005 | 0.000* |
| Biochemical | |||||
| Triglyceride (mmol/L) | 0.85 ± 0.22 | 0.93 ± 0.34 | 0.08 ± 0.24 | 1.360 | 0.193 |
| Total Cholesterol (mmol/L) | 3.93 ± 0.71 | 3.66 ± 0.60 | -0.27 ± 0.54 | -2.032 | 0.059 |
| High Density Lipoprotein (mmol/L) | 1.28 ± 0.37 | 1.25 ± 0.30 | -0.03 ± 0.18 | -0.705 | 0.491 |
| Low Density Lipoprotein (mmol/L) | 2.14 ± 0.64 | 1.94 ± 0.62 | -0.20 ± 0.64 | -1.297 | 0.213 |
| Oral Glucose Tolerance Test | |||||
| Fasting Blood Glucose (mmol/L) | 4.87 ± 0.40 | 4.90 ± 0.38 | 0.03 ± 0.57 | 0.214 | 0.833 |
| Blood Glucose 30 minutes (mmol/L) | 8.58 ± 1.58 | 8.34 ± 2.20 | -0.24 ± 2.38 | -0.417 | 0.682 |
| Blood Glucose 60 minutes (mmol/L) | 7.27 ± 1.34 | 7.65 ± 1.34 | 0.38 ± 1.50 | 1.049 | 0.310 |
| Blood Glucose 90 minutes (mmol/L) | 6.72 ± 0.82 | 6.62 ± 0.77 | -0.09 ± 1.22 | -0.317 | 0.755 |
| Blood Glucose 120 minutes (mmol/L) | 5.93 ± 0.78 | 5.62 ± 1.04 | -0.31 ± 1.00 | -1.282 | 0.218 |
| Area Under Curve (mmol/L.min) | 14.00 ± 1.25 | 14.20 ± 1.39 | 0.21 ± 1.02 | 0.835 | 0.416 |
| Fat oxidation | |||||
| Resting Fat Oxidation (gmin-1) | 0.08 ± 0.03 | 0.09 ± 0.03 | 0.01 ± 0.04 | 1.077 | 0.297 |
| Fat Oxidation at 20% VO2max (gmin-1) | 0.25 ± 0.06 | 0.26 ± 0.06 | 0.01 ± 0.06 | 0.763 | 0.456 |
| Fat Oxidation at 30% VO2max (gmin-1) | 0.24 ± 0.07 | 0.26 ± 0.06 | 0.02 ± 0.06 | 1.392 | 0.183 |
| Fat Oxidation at 40% VO2max (gmin-1) | 0.23 ± 0.10 | 0.29 ± 0.07 | 0.05 ± 0.09 | 2.423 | 0.028* |
| Fat Oxidation at 50% VO2max (gmin-1) | 0.19 ± 0.08 | 0.28 ± 0.11 | 0.09 ± 0.09 | 4.446 | 0.000* |
| Fat Oxidation at 60% VO2max (gmin-1) | 0.14 ± 0.11 | 0.23 ± 0.10 | 0.08 ± 0.12 | 2.891 | 0.011* |
| Fat Oxidation at 70% VO2max (gmin-1) | 0.09 ± 0.09 | 0.16 ± 0.11 | 0.07 ± 0.10 | 2.960 | 0.009* |
| Data are shown as mean ± SD; * p<0.05 | |||||
ACE I/D Genotype and Response to Exercise Training
Dependent variables responses among ACE genotype groups at pre-test and post-test are shown in Table 5. A 3 (group) × 2 (time) ANOVA with repeated measures on the anthropometric, cardiovascular, and biochemical variables demonstrated no interaction between ACE I/D gene polymorphism and 4 weeks of endurance training on the anthropometric (F(12, 18)=1.195, p=0.356, Wilks’ Λ=0.310), cardiovascular (F(12, 18)=0.930, p=0.540, Wilks’ Λ=0.381), and biochemical (F(8, 22)=1.195, p=0.346, Wilks’ Λ=0.486) variables responses following the training program. OGTT value also did not differ by ACE I/D genotype (F(12, 18)=0.715, p=0.719, Wilks’ Λ=0.458).
| Variables | Genotype | Pre-Training | Post-Training | Interaction (genotype × training session) | Training effect (within-subject) | Genotype effect (between subjects) |
|---|---|---|---|---|---|---|
| Anthropometric | ||||||
| Body Weight (kg) | II | 73.93 ± 5.37 | 75.97 ± 1.40 | 0.314 | 0.567 | 0.825 |
| ID | 74.22 ± 14.88 | 74.02 ± 15.59 | ||||
| DD | 72.97 ± 12.42 | 67.52 ± 17.27 | ||||
| Body Mass Index (kg/m2) | II | 23.30 ± 2.91 | 23.89 ± 1.75 | 0.322 | 0.532 | 0.945 |
| ID | 23.88 ± 2.43 | 23.79 ± 2.61 | ||||
| DD | 24.12 ± 3.50 | 22.36 ± 5.39 | ||||
| Waist (cm) | II | 80.70 ± 5.28 | 81.77 ± 2.54 | 0.424 | 0.682 | 0.930 |
| ID | 79.90 ± 8.82 | 78.46 ± 9.13 | ||||
| DD | 79.20 ± 8.96 | 78.69 ± 11.52 | ||||
| Hip Circumference (cm) | II | 96.77 ± 2.20 | 97.77 ± 0.47 | 0.092 | 0.326 | 0.753 |
| ID | 98.89 ± 5.65 | 98.64 ± 6.05 | ||||
| DD | 97.70 ± 7.21 | 94.77 ± 8.25 | ||||
| Waist Hip Ratio | II | 0.83 ± 0.04 | 0.84 ± 0.03 | 0.082 | 0.657 | 0.549 |
| ID | 0.81 ± 0.05 | 0.79 ± 0.05 | ||||
| DD | 0.81 ± 0.04 | 0.83 ± 0.06 | ||||
| Body Fat (%) | II | 16.50 ± 5.14 | 17.93 ± 7.54 | 0.689 | 0.461 | 0.827 |
| ID | 20.30 ± 6.86 | 20.69 ± 7.49 | ||||
| DD | 19.36 ± 8.81 | 19.16 ± 8.74 | ||||
| Cardiovascular | ||||||
| Systolic Blood Pressure (mmHg) | II | 119.67 ± 10.41 | 117.67 ± 10.69 | 0.888 | 0.326 | 0.786 |
| ID | 120.86 ± 4.45 | 120.29 ± 5.71 | ||||
| DD | 119.43 ± 7.00 | 117.57 ± 4.58 | ||||
| Diastolic Blood Pressure (mmHg) | II | 74.67 ± 4.93 | 72.67 ± 5.03 | 0.724 | 0.643 | 0.776 |
| ID | 73.14 ± 6.09 | 76.14 ± 7.45 | ||||
| DD | 75.00 ± 9.45 | 77.43 ± 6.93 | ||||
| Mean Arterial Pressure (mmHg) | II | 89.67 ± 6.74 | 87.67 ± 4.73 | 0.701 | 0.877 | 0.886 |
| ID | 89.05 ± 3.76 | 90.86 ± 5.88 | ||||
| DD | 89.81 ± 7.53 | 90.81 ± 5.57 | ||||
| Resting Heart Rate (beat per minute) | II | 50.96 ± 9.87 | 52.67 ± 11.24 | 0.533 | 0.463 | 0.625 |
| ID | 57.38 ± 6.01 | 55.11 ± 8.96 | ||||
| DD | 54.55 ± 8.13 | 51.45 ± 7.59 | ||||
| Resting Pulse Pressure (mmHg) | II | 6037.67 ± 707.84 | 6184.33 ± 1282.96 | 0.565 | 0.341 | 0.460 |
| ID | 6934.82 ± 782.38 | 6622.09 ± 1082.04 | ||||
| DD | 6520.43 ± 1098.19 | 6044.09 ± 881.73 | ||||
| Maximal Oxygen Consumption (L/min) | II | 3.48 ± 0.47 | 3.83 ± 0.21 | 0.532 | 0.003* | 0.594 |
| ID | 3.07 ± 0.86 | 3.26 ± 0.95 | ||||
| DD | 3.14 ± 0.57 | 3.30 ± 0.59 | ||||
| Peak Power Output (Watts) | II | 302.67 ± 45.39 | 333.33 ± 26.56 | 0.642 | 0.000* | 0.516 |
| ID | 264.00 ± 79.67 | 284.00 ± 82.06 | ||||
| DD | 256.00 ± 46.43 | 284.00 ± 44.20 | ||||
| Biochemical | ||||||
| Triglyceride (mmol/L) | II | 0.93 ± 0.27 | 0.75 ± 0.15 | 0.081 | 0.618 | 0.561 |
| ID | 0.74 ± 0.12 | 0.91 ± 0.27 | ||||
| DD | 0.93 ± 0.24 | 1.02 ± 0.45 | ||||
| Total Cholesterol (mmol/L) | II | 3.28 ± 0.54 | 3.36 ± 0.50 | 0.418 | 0.180 | 0.110 |
| ID | 4.25 ± 0.42 | 4.00 ± 0.46 | ||||
| DD | 3.89 ± 0.85 | 3.45 ± 0.66 | ||||
| High Density Lipoprotein (mmol/L) | II | 1.14 ± 0.27 | 1.06 ± 0.23 | 0.476 | 0.404 | 0.247 |
| ID | 1.45 ± 0.38 | 1.38 ± 0.25 | ||||
| DD | 1.15 ± 0.38 | 1.19 ± 0.33 | ||||
| Low Density Lipoprotein (mmol/L) | II | 1.73 ± 0.64 | 1.80 ± 0.70 | 0.259 | 0.372 | 0.493 |
| ID | 2.22 ± 0.68 | 2.21 ± 0.67 | ||||
| DD | 2.25 ± 0.63 | 1.74 ± 0.53 | ||||
| Oral Glucose Tolerance Test | ||||||
| Fasting Blood Glucose (mmol/L) | II | 4.60 ± 0.72 | 4.97 ± 0.15 | 0.544 | 0.547 | 0.799 |
| ID | 4.94 ± 0.36 | 4.87 ± 0.48 | ||||
| DD | 4.91 ± 0.29 | 4.90 ± 0.38 | ||||
| Blood Glucose 30 minutes (mmol/L) | II | 8.83 ± 1.91 | 9.53 ± 1.96 | 0.715 | 0.926 | 0.157 |
| ID | 7.99 ± 1.20 | 7.27 ± 2.67 | ||||
| DD | 9.07 ± 1.80 | 8.90 ± 1.44 | ||||
| Blood Glucose 60 minutes (mmol/L) | II | 8.43 ± 1.76 | 7.70 ± 1.04 | 0.373 | 0.672 | 0.479 |
| ID | 6.87 ± 1.22 | 7.37 ± 1.31 | ||||
| DD | 7.17 ± 1.17 | 7.91 ± 1.59 | ||||
| Blood Glucose 90 minutes (mmol/L) | II | 7.37 ± 1.00 | 6.53 ± 1.27 | 0.517 | 0.484 | 0.553 |
| ID | 6.57 ± 0.96 | 6.53 ± 0.78 | ||||
| DD | 6.59 ± 0.51 | 6.76 ± 0.64 | ||||
| Blood Glucose 120 minutes (mmol/L) | II | 5.23 ± 0.15 | 5.10 ± 0.62 | 0.699 | 0.328 | 0.248 |
| ID | 6.03 ± 0.66 | 5.46 ± 1.04 | ||||
| DD | 6.13 ± 0.94 | 6.00 ± 1.14 | ||||
| Area Under Curve (mmol/L.min) | II | 14.78 ± 1.65 | 14.40 ± 1.93 | 0.582 | 0.733 | 0.452 |
| ID | 13.49 ± 1.33 | 13.81 ± 1.21 | ||||
| DD | 14.18 ± 0.93 | 14.51 ± 1.46 | ||||
| Fat oxidation | ||||||
| Resting Fat Oxidation (gmin-1) | II | 0.08 ± 0.05 | 0.12 ± 0.01 | 0.348 | 0.162 | 0.138 |
| ID | 0.07 ± 0.04 | 0.06 ± 0.03 | ||||
| DD | 0.08 ± 0.03 | 0.09 ± 0.03 | ||||
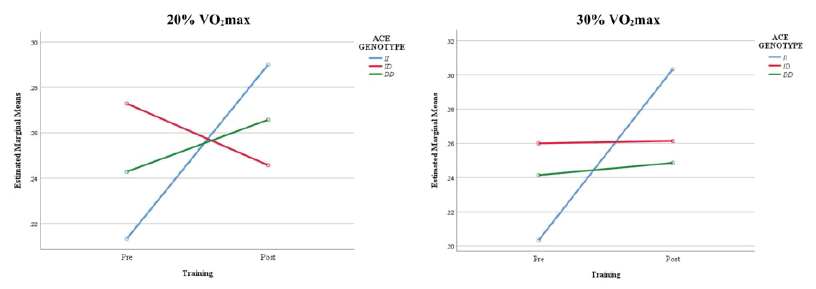
| Fat Oxidation at 20% VO2max (gmin-1) | II | 0.21 ± 0.06 | 0.29 ± 0.06 | 0.038* | 0.109 | 0.972 |
| ID | 0.27 ± 0.07 | 0.25 ± 0.07 | ||||
| DD | 0.24 ± 0.04 | 0.27 ± 0.05 | ||||
| Fat Oxidation at 30% VO2max (gmin-1) | II | 0.20 ± 0.05 | 0.30 ± 0.04 | 0.043* | 0.022* | 0.896 |
| ID | 0.26 ± 0.08 | 0.26 ± 0.08 | ||||
| DD | 0.24 ± 0.08 | 0.25 ± 0.04 | ||||
| Fat Oxidation at 40% VO2max (gmin-1) | II | 0.20 ± 0.07 | 0.33 ± 0.04 | 0.263 | 0.011* | 0.85 |
| ID | 0.26 ± 0.10 | 0.28 ± 0.09 | ||||
| DD | 0.22 ± 0.12 | 0.28 ± 0.06 | ||||
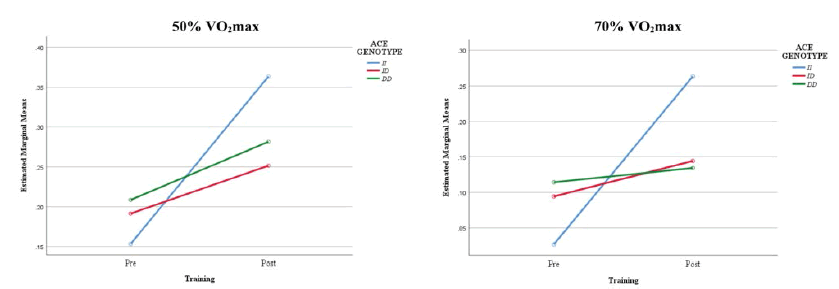
| Fat Oxidation at 50% VO2max (gmin-1) | II | 0.15 ± 0.05 | 0.36 ± 0.06 | 0.016* | 0.000* | 0.806 |
| ID | 0.19 ± 0.07 | 0.25 ± 0.10 | ||||
| DD | 0.21 ± 0.10 | 0.28 ± 0.12 | ||||
| Fat Oxidation at 60% VO2max (gmin-1) | II | 0.12 ± 0.10 | 0.32 ± 0.03 | 0.207 | 0.003* | 0.669 |
| ID | 0.16 ± 0.10 | 0.23 ± 0.11 | ||||
| DD | 0.14 ± 0.13 | 0.19 ± 0.10 | ||||
| Fat Oxidation at 70% VO2max (gmin-1) | II | 0.03 ± 0.03 | 0.26 ± 0.01 | 0.000* | 0.000* | 0.921 |
| ID | 0.09 ± 0.07 | 0.14 ± 0.10 | ||||
| DD | 0.11 ± 0.12 | 0.13 ± 0.12 | ||||
| Note: Data are shown as mean ± SD; * p<0.05 | ||||||
However, there was a statistically significant interaction effect between ACE I/D genotype and 4 weeks of endurance training on the fat oxidation variables (F(14, 16)=3.269, p=0.013; Wilks’ Λ=0.067). Figure 1 described the estimated marginal means of the interaction effect between ACE I/D genotype and endurance training on fat oxidation at 20% and 30% VO2 max. The interaction observed for the scores at 20% and 30% VO2 max indicated that although the ID genotype group had much higher fat oxidation rates at 20% and 30% VO2 max than the other genotype groups before the endurance training, the II genotype group obtained the highest rates after the training program.
Figure 2 illustrated the estimated marginal means of the interaction effect between ACE I/D genotype and endurance training on fat oxidation at 50% and 70% VO2 max. The interaction indicated that although the DD genotype group had much higher fat oxidation scores at 50% and 70% VO2 max than the other genotype groups before the endurance training, the II genotype group obtained the highest scores after the training program.
Discussion
This study aimed to determine whether ACE I/D gene polymorphism is associated with an adaptive response to shortterm endurance training. While previous studies demonstrated the association of ACE I/D gene polymorphism on the enhancement in endurance performance following the endurance training among trained individuals, the information on the association between ACE I/D gene polymorphism and such parameter was not well established for untrained individuals [1,2,4,5]. It is important to note that this information is important as it reduces the gene-environment interaction such as the training effect of the trained individuals.
The present study found significant differences in VO2 max, peak power output, and fat oxidation at (40%, 50%, 60%, and 70%) VO2 max between the pre-training and post-training, which showed that 4 week of endurance training program elicits improvements in endurance performances of untrained adults. Interestingly, greater improvements in fat oxidation rate scores after the training program were more pronounced among participants with II genotype compared to other genotypes. This result appears to reflect the suggestion in the previous study that the II genotype can be correlated with enhanced medium-long aerobic endurance performance, while the DD genotype appears to be better for enhancing performance over the shorter duration and greater intensity durability [2].
The baseline data obtained from the present study suggest that in overall participants comprised of both genders, the possession of certain ACE I/D genotypes on studied parameters is not obvious, except for the HDL variable. However, when the baseline data were analyzed by gender, we found that the waist-hip ratio in females was significantly lower in the ID genotype group compared to other genotype groups. Meanwhile, DBP, MAP, and total cholesterol in the male may be related to the ACE I/D gene polymorphism as DBP and MAP were significantly lower in the ID genotype group compared to another genotype group. Meanwhile, the total cholesterol was significantly lowered among II genotype carriers. Although studies on the sex differences in fat oxidation during exercise suggest that there is variability in fat oxidation owing to sex exist due to the inherent hormonal differences specific to men and women, the study present showed a lack of association of baseline OGTT-based ACE I/D gene polymorphism with fat oxidation values regardless of gender [25-27]. This result extends the findings from Huang, et al., who have found no significant association of the ACE I/D polymorphism with glucose tolerance among non-diabetic individuals [12]. One possible explanation for the lacking association observed in the present study may also be due that our subjects are healthy individuals. The association between ACE I/D polymorphism and glucose tolerance can be observed in high-risk individuals such as diabetic patients or overweight individuals as observed in another study [12].
Although there was a small association found between the studied gene variant with the baseline parameters, the present study observed that individual metabolic responses to endurance training were heterogeneous. Surprisingly, II genotype carriers who showed a lower baseline in fat oxidation rate have a better response to endurance training compared to other genotype groups who had higher baseline values. This finding was consistent with previous studies that have attempted to identify if possession of the I allele would enhance the effect of exercise training on training adaptation [28]. In comparison with the D allele carrier, previous studies demonstrated that the I allele carrier was shown to enhance mechanical efficiency in trained muscle following an 11-week program of aerobic training, increased aortic distensibility due to chronic prolonged training, greater exercise adherence to 6-month exercise training at 60% and 85% of maximal exercise capacity, as well as higher improvement in 30 minutes (at 70% of heart rate reserve) running speed performance after a 6-week anaerobic and aerobic training program [1,28-30].
Following the above-mentioned findings, we can assume that there was a greater physiological adaptation among II genotype individuals compared to other genotypes for aerobic training. The mechanisms of how ACE I/D gene polymorphism influences fat oxidation response to endurance training are unclear and not well reported. The endocrine system is reported to be responsible for the regulation of fat oxidation at rest and during exercise [31]. The activation of the hormone system during exercises such as the Renin-Angiotensin-Aldosterone System (RAAS) and its component; angiotensin II levels which are determined by the ACE I/D gene polymorphism could be a potential mechanism that would permit the II genotype individuals to perform in endurance activities better than those with the DD genotype. It is speculated that during exercise, the ACE level which was reported lower in individuals with II genotype will make the conversion of ANG I to ANG II decreased, leading to a reduction in skeletal muscle vasoconstriction, and an increase in blood flow oxygenated to the working muscles [32,33].
The low level of ACE in RAS is also thought will enhance fat oxidation by other indirect mechanisms depending on the specific tissues it stimulates, the level of activation, or the physiological state of the human. A current study reported that the ACE modulated expression pathway being activated by endurance exercise is associated with carbohydrate and lipid metabolism in skeletal muscle [34]. Another important determinant of endurance performance, strongly involved in the resistance to fatigue and adaptation to endurance exercise, is substrate utilization. Substrate utilization differs according to muscle phenotype [7]. The soleus, an oxidative muscle, can actively oxidize fatty acids, whereas the gastrocnemius, a predominantly glycolytic muscle, produces energy mainly from glucose use [7]. Although the majority of studies have focused on the systemic influences of the RAS on metabolism, there is also evidence that brain angiotensin may also have a determining influence [35]. A study showed that rats with a deficiency in brain angiotensinogen that is induced by an angiotensinogen-antisense gene coupled to a glial cell-specific promoter were found to have reduced body fat composition, and improved glucose tolerance, low blood pressure, and increased appetite compared with wild-type rats [35]. However, these components were not evaluated in this study which indicates that more work is thus needed to assess this potential mechanism for the adaptive response to endurance training.
Contradictory findings for associations with the ACE I/D gene polymorphism may also be the result of genetic effects interacting with specific types of training. The presence of the D allele is associated most strongly with the shortduration, power-oriented events, whereas the I allele has been associated with performance in longer-duration events [24]. The potentially favorable effect of possession of the D allele on short-duration, power-oriented events could be due to the production of ANG II in the skeletal muscle. A greater local ANG II production in the skeletal muscle has been reported to increase protein synthesis and cell hypertrophy, thereby inducing muscle contraction for maximal power [36,37]. Following this evidence, as this study employed endurance training, the greater adaptive response observed among II genotype individuals might be interpreted due to reduced ACE serum levels during exercise. The current study, however, does not measure the ACE serum which requires future research to validate this potential mechanism.
Despite all positive findings, the mixed and non-significant results for other parameters in our study were speculated may be generally due to the small number of participants examined in this study. Our sample group consisted of healthy individuals and involved mixed-gender participants. For the training program, we include both genders as the number of females is low, and it makes it impossible to compare it by genotype. Therefore, it remains unknown if the effects of the ACE I/D polymorphism on training adaptation may differ by gender. The responses to training may easily be observed among special populations such as diabetes and cardiovascular patient. The number of training sessions may also appear to contribute to this controversial finding. Given that the absence of a concrete guideline on the number of training sessions needs to be conducted to elucidate the effects of the ACE I/D gene polymorphism on training response, this speculation needs further investigation.
Even with these limitations, the findings currently available are reasonably reliable since each research participant is comparatively uniform in terms of health and training status. The genetic evaluation was reliable and unbiased because the genotype distribution was in the Hardy-Weinberg balance. The intervention was standardized by participants with the same trained investigator who conducted the training intervention. The participants underwent familiarisation procedures to reduce the variability in starting point measurements caused by apprehension. This study was conducted as a double-blind study, in which the participant and a qualified researcher who conducted the data collection did not know the genotype of the participants.
Conclusion
In summary, this research endorses the idea that the ACE I/D polymorphism correlated with adaptation to short-term endurance training. The individuals possessed of II genotype are more favorable to endurance training compared to other genotype carriers. This finding shows that before an individual engages in a certain exercise program for health management, genetic testing is essential to ensure that efficiency and the benefits of the exercise training program are fully utilized. More investigation with larger sample size and longer duration of exercise program is warranted to delineate and confirm the current findings.
Declarations
Conflict of Interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Cam, S., et al. "ACE I/D gene polymorphism and aerobic endurance development in response to training in a non-elite female cohort." Journal of Sports Medicine and Physical Fitness, Vol. 47, No. 2, 2007, pp. 234-38.
Google Scholar - Montgomery, H. E., et al. "Human gene for physical performance." Nature, Vol. 393, No. 6682, 1998, pp. 221-22.
Google Scholar Crossref - Sonna, Larry A., et al. "Angiotensin-converting enzyme genotype and physical performance during US Army basic training." Journal of Applied Physiology, Vol. 91, No. 3, 2001, pp. 1355-63.
Google Scholar Crossref - Vaughan, David, et al. "The angiotensin converting enzyme insertion/deletion polymorphism alters the response of muscle energy supply lines to exercise." European Journal of Applied Physiology, Vol. 113, No. 7, 2013, pp. 1719-29.
Google Scholar Crossref - Woods, David R., and Hugh E. Montgomery. "Angiotensin-converting enzyme and genetics at high altitude." High Altitude Medicine & Biology, Vol. 2, No. 2, 2001, pp. 201-10.
Google Scholar Crossref - Woods, D., et al. "Endurance enhancement related to the human angiotensin I-converting enzyme ID polymorphism is not due to differences in the cardiorespiratory response to training." European Journal of Applied Physiology, Vol. 86, No. 3, 2002, pp. 240-44.
Google Scholar Crossref - Bahi, Lahoucine, et al. "Does ACE inhibition enhance endurance performance and muscle energy metabolism in rats?" Journal of Applied Physiology, Vol. 96, No. 1, 2004, pp. 59-64.
Google Scholar Crossref - Van Dyk, D. J., et al. "Increased serum angiotensin converting enzyme activity in type T insulin-dependent diabetes mellitus: Its relation to metabolic control and diabetic complications." European Journal of Clinical Investigation, Vol. 24, No. 7, 1994, pp. 463-67.
Google Scholar Crossref - Fiatal, Szilvia, et al. "Insertion/deletion polymorphism of angiotensin-1 converting enzyme is associated with metabolic syndrome in Hungarian adults." Journal of the Renin-Angiotensin-Aldosterone System, Vol. 12, No. 4, 2011, pp. 531-38.
Google Scholar Crossref - Hagberg, James M., et al. "Exercise training-induced blood pressure and plasma lipid improvements in hypertensives may be genotype dependent." Hypertension, Vol. 34, No. 1, 1999, pp. 18-23.
Google Scholar Crossref - Bassuk, Shari S., and JoAnn E. Manson. "Epidemiological evidence for the role of physical activity in reducing risk of type 2 diabetes and cardiovascular disease." Journal of Applied Physiology, Vol. 99, No. 3, 2005, pp. 1193-204.
Google Scholar Crossref - Huang, Xiao-Hong, et al. "Relationship of the angiotensin-converting enzyme gene polymorphism to glucose intolerance, insulin resistance, and hypertension in NIDDM." Human Genetics, Vol. 102, No. 3, 1998, pp. 372-78.
Google Scholar Crossref - Ng, D. P. K., et al. "Angiotensin-I converting enzyme insertion/deletion polymorphism and its association with diabetic nephropathy: A meta-analysis of studies reported between 1994 and 2004 and comprising 14,727 subjects." Diabetologia, Vol. 48, No. 5, 2005, pp. 1008-16.
Google Scholar Crossref - Rogers, Marc A., et al. "Improvement in glucose tolerance after 1 wk of exercise in patients with mild NIDDM." Diabetes Care, Vol. 11, No. 8, 1988, pp. 613-18.
Google Scholar Crossref - Smutok, M. A., et al. "Effects of exercise training modality on glucose tolerance in men with abnormal glucose regulation." International Journal of Sports Medicine, Vol. 15, No. 06, 1994, pp. 283-89.
Google Scholar - Snowling, Neil J., and Will G. Hopkins. "Effects of different modes of exercise training on glucose control and risk factors for complications in type 2 diabetic patients: A meta-analysis." Diabetes Care, Vol. 29, No. 11, 2006, pp. 2518-27.
Google Scholar Crossref - Wilmore, Jack H., et al. "Relationship of changes in maximal and submaximal aerobic fitness to changes in cardiovascular disease and non [ndash] insulin-dependent diabetes mellitus risk factors with endurance training: The HERITAGE Family Study." Metabolism-Clinical and Experimental, Vol. 50, No. 11, 2001, pp. 1255-63.
Google Scholar Crossref - Lieberman, Jack, and Adriana Sastre. "Serum angiotensin-converting enzyme: Elevations in diabetes mellitus." Annals of Internal Medicine, Vol. 93, No. 6, 1980, pp. 825-26.
Google Scholar Crossref - Dengel, Donald R., et al. "Exercise-induced changes in insulin action are associated with ACE gene polymorphisms in older adults." Physiological Genomics, Vol. 11, No. 2, 2002, pp. 73-80.
Google Scholar Crossref - Hurlbut, D. E., et al. "Does age, sex, or ACE genotype affect glucose and insulin responses to strength training?" Journal of Applied Physiology, Vol. 92, No. 2, 2002, pp. 643-50.
Google Scholar Crossref - Luptakova, Lenka, et al. "Association of CILP2 and ACE gene polymorphisms with cardiovascular risk factors in Slovak midlife women." BioMed Research International, Vol. 2013, 2013.
Google Scholar - Reardon, Trent F., et al. "Creatine supplementation does not enhance submaximal aerobic training adaptations in healthy young men and women." European Journal of Applied Physiology, Vol. 98, No. 3, 2006, pp. 234-41.
Google Scholar Crossref - World Health Organization & International Diabetes Federation. "Definition and diagnosis of diabetes mellitus and intermediate hyperglycaemia: Report of a WHO/IDF consultation." World Health Organization, 2006.
Google Scholar - Puthucheary, Zudin, et al. "The ACE gene and human performance." Sports Medicine, Vol. 41, No. 6, 2011, pp. 433-48.
Google Scholar Crossref - Isacco, Laurie, Pascale Duche, and Nathalie Boisseau. "Influence of hormonal status on substrate utilization at rest and during exercise in the female population." Sports Medicine, Vol. 42, No. 4, 2012, pp. 327-42.
Google Scholar Crossref - Maher, Amy C., et al. "Women have higher protein content of β-oxidation enzymes in skeletal muscle than men." PLoS One, Vol. 5, No. 8, 2010, p. e12025.
Google Scholar Crossref - Varlamov, Oleg, Cynthia L. Bethea, and Charles T. Roberts Jr. "Sex-specific differences in lipid and glucose metabolism." Frontiers in Endocrinology, Vol. 5, 2015, p. 241.
Google Scholar Crossref - Williams, Alun G., et al. "Circulating angiotensin converting enzyme activity is correlated with muscle strength." Medicine & Science in Sports & Exercise, Vol. 37, No. 6, 2005, pp. 944-48.
Google Scholar Crossref - Tanrıverdi, Halil, et al. "Effects of angiotensin-converting enzyme polymorphism on aortic elastic parameters in athletes." Cardiology, Vol. 104, No. 3, 2005, pp. 113-19.
Google Scholar Crossref - Thompson, Paul D., et al. "Angiotensin‐converting enzyme genotype and adherence to aerobic exercise training." Preventive Cardiology, Vol. 9, No. 1, 2006, pp. 21-24.
Google Scholar Crossref - Horowitz, Jeffrey F., and Samuel Klein. "Lipid metabolism during endurance exercise." The American Journal of Clinical Nutrition, Vol. 72, No. 2, 2000, pp. 558S-63S.
Google Scholar Crossref - Rigat, Brigitte, et al. "An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels." The Journal of Clinical Investigation, Vol. 86, No. 4, 1990, pp. 1343-46.
Google Scholar Crossref - Sayed-Tabatabaei, F. A., et al. "ACE polymorphisms." Circulation Research, Vol. 98, No. 9, 2006, pp. 1123-33.
Google Scholar Crossref - Valdivieso, Paola, et al. "The metabolic response of skeletal muscle to endurance exercise is modified by the ACE-I/D gene polymorphism and training state." Frontiers in Physiology, Vol. 8, 2017, p. 993.
Google Scholar Crossref - Jayasooriya, Anura P., et al. "Mice lacking angiotensin-converting enzyme have increased energy expenditure, with reduced fat mass and improved glucose clearance." Proceedings of the National Academy of Sciences, Vol. 105, No. 18, 2008, pp. 6531-36.
Google Scholar Crossref - Jones, Alun, and David R. Woods. "Skeletal muscle RAS and exercise performance." The International Journal of Biochemistry & Cell Biology, Vol. 35, No. 6, 2003, pp. 855-66.
Google Scholar Crossref - Rattigan, S. T. E. P. H. E. N., et al. "Perfused skeletal muscle contraction and metabolism improved by angiotensin II-mediated vasoconstriction." American Journal of Physiology-Endocrinology and Metabolism, Vol. 271, No. 1, 1996, pp. E96-E103.
Google Scholar Crossref